+Search query
-Structure paper
| Title | Insights into the Activation Mechanism of HCA1, HCA2, and HCA3. |
|---|---|
| Journal, issue, pages | J Med Chem, Vol. 68, Issue 4, Page 4527-4539, Year 2025 |
| Publish date | Feb 27, 2025 |
 Authors Authors | Jiening Wang / Yuxia Qian / Zhen Han / Yize Wang / Yanru Liu / Jie Li / Qingmiao Duanmu / Sheng Ye / Anna Qiao / Shan Wu /  |
| PubMed Abstract | Hydroxy-carboxylic acid receptors HCA1, HCA2, and HCA3 can be activated by important intermediates of energy metabolism. Despite the research focusing on HCA2, its clinical application has been ...Hydroxy-carboxylic acid receptors HCA1, HCA2, and HCA3 can be activated by important intermediates of energy metabolism. Despite the research focusing on HCA2, its clinical application has been limited by adverse effects. Therefore, the role of HCA1 as a promising target for the treatment of lipolysis warrants further exploration. As HCAs exhibit high similarity when activated with diverse selective agonists, a conserved yet unique activation mechanism for HCAs remains undisclosed. Herein, we unveil the cryo-electron microscopy structures of the 3,5-DHBA-HCA1-Gi signaling complex, the acifran- and MK6892-bound HCA2-Gi signaling complexes, and the acifran-HCA3-Gi signaling complex. Comparative analysis across HCAs reveals key residues in HCA1 contributing to the stabilization of the ligand-binding pocket. Furthermore, chimeric complexes and mutational analyses identify residues that are pivotal for HCA2 and HCA3 selectivity. Our findings elucidate critical structural insights into the mechanisms of ligand recognition and activation within HCA1 and broaden our comprehension of ligand specificity binding across the HCA family. |
 External links External links |  J Med Chem / J Med Chem /  PubMed:39936872 / PubMed:39936872 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.01 Å |
| Structure data | EMDB-62557, PDB-9kt6: EMDB-62558, PDB-9kt7: EMDB-62559, PDB-9kt8: EMDB-62560, PDB-9kt9: |
| Chemicals | 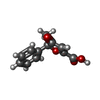 ChemComp-P9X:  ChemComp-FI7: 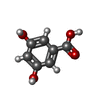 ChemComp-34D: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / cryo-EM; G-protein-coupled receptors; Hydroxycarboxylic acid receptors / SIGNALING PROTEIN/IMMUNE SYSTEM / SIGNALING PROTEIN-IMMUNE SYSTEM complex / G-protein-coupled receptors; Hydroxycarboxylic acid receptors / cryo-EM; SIGNALING PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)