+Search query
-Structure paper
| Title | Structure of blood cell-specific tubulin and demonstration of dimer spacing compaction in a single protofilament. |
|---|---|
| Journal, issue, pages | J Biol Chem, Vol. 301, Issue 2, Page 108132, Year 2025 |
| Publish date | Dec 24, 2024 |
 Authors Authors | Felipe Montecinos / Elif Eren / Norman R Watts / Dan L Sackett / Paul T Wingfield /  |
| PubMed Abstract | Microtubule (MT) function plasticity originates from its composition of α- and β-tubulin isotypes and the posttranslational modifications of both subunits. Aspects such as MT assembly dynamics, ...Microtubule (MT) function plasticity originates from its composition of α- and β-tubulin isotypes and the posttranslational modifications of both subunits. Aspects such as MT assembly dynamics, structure, and anticancer drug binding can be modulated by αβ-tubulin heterogeneity. However, the exact molecular mechanism regulating these aspects is only partially understood. A recent insight is the discovery of expansion and compaction of the MT lattice, which can occur via fine modulation of dimer longitudinal spacing mediated by GTP hydrolysis, taxol binding, protein binding, or isotype composition. Here, we report the first structure of the blood cell-specific α1/β1-tubulin isolated from the marginal band of chicken erythrocytes (ChET) determined to a resolution of 3.2 Å by cryo-EM. We show that ChET rings induced with cryptophycin-52 (Cp-52) are smaller in diameter than HeLa cell line tubulin (HeLaT) rings induced with Cp-52 and composed of the same number of heterodimers. We observe compacted interdimer and intradimer curved protofilament interfaces, characterized by shorter distances between ChET subunits and accompanied by conformational changes in the β-tubulin subunit. The compacted ChET interdimer interface brings more residues near the Cp-52 binding site. We measured the Cp-52 apparent binding affinities of ChET and HeLaT by mass photometry, observing small differences, and detected the intermediates of the ring assembly reaction. These findings demonstrate that compaction/expansion of dimer spacing can occur in a single protofilament context and that the subtle structural differences between tubulin isotypes can modulate tubulin small molecule binding. |
 External links External links |  J Biol Chem / J Biol Chem /  PubMed:39725029 / PubMed:39725029 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 - 3.52 Å |
| Structure data | EMDB-45263, PDB-9c6r: EMDB-45265, PDB-9c6s: |
| Chemicals |  ChemComp-GTP: 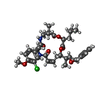 ChemComp-YGY:  ChemComp-GDP: |
| Source |
|
 Keywords Keywords | CELL CYCLE / Tubulin Beta-6 chain / TBB6_CHICK / TUBB1 / Ring C8 / Hematopoietic isotype Cryptophycin / Anticancer / GTPase / Cytoskeleton / Cryptophycin-52 / Hematopoietic / Beta-6 tubulin chain / Class VI |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








