+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Blood cell-specific tubulin in complex with Cryptophycin-52 | |||||||||
 Map data Map data | post-processed cryo-em map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tubulin Beta-6 chain / TBB6_CHICK / TUBB1 / Ring C8 / Hematopoietic isotype Cryptophycin / Anticancer / GTPase / Cytoskeleton / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationIntraflagellar transport / Kinesins / COPI-dependent Golgi-to-ER retrograde traffic / Aggrephagy / Resolution of Sister Chromatid Cohesion / EML4 and NUDC in mitotic spindle formation / Separation of Sister Chromatids / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport ...Intraflagellar transport / Kinesins / COPI-dependent Golgi-to-ER retrograde traffic / Aggrephagy / Resolution of Sister Chromatid Cohesion / EML4 and NUDC in mitotic spindle formation / Separation of Sister Chromatids / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / structural constituent of cytoskeleton / microtubule cytoskeleton organization / mitotic cell cycle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / GTPase activity / GTP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Montecinos F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2025 Journal: J Biol Chem / Year: 2025Title: Structure of blood cell-specific tubulin and demonstration of dimer spacing compaction in a single protofilament. Authors: Felipe Montecinos / Elif Eren / Norman R Watts / Dan L Sackett / Paul T Wingfield /  Abstract: Microtubule (MT) function plasticity originates from its composition of α- and β-tubulin isotypes and the posttranslational modifications of both subunits. Aspects such as MT assembly dynamics, ...Microtubule (MT) function plasticity originates from its composition of α- and β-tubulin isotypes and the posttranslational modifications of both subunits. Aspects such as MT assembly dynamics, structure, and anticancer drug binding can be modulated by αβ-tubulin heterogeneity. However, the exact molecular mechanism regulating these aspects is only partially understood. A recent insight is the discovery of expansion and compaction of the MT lattice, which can occur via fine modulation of dimer longitudinal spacing mediated by GTP hydrolysis, taxol binding, protein binding, or isotype composition. Here, we report the first structure of the blood cell-specific α1/β1-tubulin isolated from the marginal band of chicken erythrocytes (ChET) determined to a resolution of 3.2 Å by cryo-EM. We show that ChET rings induced with cryptophycin-52 (Cp-52) are smaller in diameter than HeLa cell line tubulin (HeLaT) rings induced with Cp-52 and composed of the same number of heterodimers. We observe compacted interdimer and intradimer curved protofilament interfaces, characterized by shorter distances between ChET subunits and accompanied by conformational changes in the β-tubulin subunit. The compacted ChET interdimer interface brings more residues near the Cp-52 binding site. We measured the Cp-52 apparent binding affinities of ChET and HeLaT by mass photometry, observing small differences, and detected the intermediates of the ring assembly reaction. These findings demonstrate that compaction/expansion of dimer spacing can occur in a single protofilament context and that the subtle structural differences between tubulin isotypes can modulate tubulin small molecule binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45263.map.gz emd_45263.map.gz | 67.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45263-v30.xml emd-45263-v30.xml emd-45263.xml emd-45263.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45263_fsc.xml emd_45263_fsc.xml | 21.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_45263.png emd_45263.png | 139 KB | ||
| Masks |  emd_45263_msk_1.map emd_45263_msk_1.map | 1.1 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-45263.cif.gz emd-45263.cif.gz | 7.2 KB | ||
| Others |  emd_45263_additional_1.map.gz emd_45263_additional_1.map.gz emd_45263_half_map_1.map.gz emd_45263_half_map_1.map.gz emd_45263_half_map_2.map.gz emd_45263_half_map_2.map.gz | 699.3 MB 1016.8 MB 1016.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45263 http://ftp.pdbj.org/pub/emdb/structures/EMD-45263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45263 | HTTPS FTP |
-Related structure data
| Related structure data |  9c6rMC  9c6sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45263.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45263.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed cryo-em map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45263_msk_1.map emd_45263_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: locally sharpened map (DeepEmhancer)
| File | emd_45263_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally sharpened map (DeepEmhancer) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map A
| File | emd_45263_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map B
| File | emd_45263_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chicken erythrocytes tubulin in complex with cryptophycin-52
| Entire | Name: Chicken erythrocytes tubulin in complex with cryptophycin-52 |
|---|---|
| Components |
|
-Supramolecule #1: Chicken erythrocytes tubulin in complex with cryptophycin-52
| Supramolecule | Name: Chicken erythrocytes tubulin in complex with cryptophycin-52 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 734 KDa |
-Macromolecule #1: Detyrosinated tubulin alpha-1A chain
| Macromolecule | Name: Detyrosinated tubulin alpha-1A chain / type: protein_or_peptide / ID: 1 Details: Tubulin alpha-1A chain TUBA1A,P02552,TBA1A_CHICK, NP_001292201.2 Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.188441 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLIGQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1A chain |
-Macromolecule #2: Tubulin beta-6 chain
| Macromolecule | Name: Tubulin beta-6 chain / type: protein_or_peptide / ID: 2 Details: Tubulin beta-6 chain, P09207, TBB6_CHICK, TUBB1, Hematopoietic beta-tubulin, Beta-tubulin class-VI Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.201469 KDa |
| Sequence | String: MREIVHLQIG QCGNQIGAKF WEVISDEHGI DIAGNYCGNA SLQLERINVY FNEAYSHKYV PRSILVDLEP GTMDSVRSSK IGPLFRPDN FIHGNSGAGN NWAKGHYTEG AELIENVMDV VRNECESCDC LQGFQLIHSL GGGTGSGMGT LLINKIREEY P DRIMNTFS ...String: MREIVHLQIG QCGNQIGAKF WEVISDEHGI DIAGNYCGNA SLQLERINVY FNEAYSHKYV PRSILVDLEP GTMDSVRSSK IGPLFRPDN FIHGNSGAGN NWAKGHYTEG AELIENVMDV VRNECESCDC LQGFQLIHSL GGGTGSGMGT LLINKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNAILS IHQLIENTDE TFCIDNEALY DICFRTLKLT NPTYGDLNHL VSLTMSGVTT SL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTARG SQQYRALSVP ELTQQMFDAR NMMAACDPRR GRYLTVACIF RGR MSTREV DEQLLSVQTK NSSYFVEWIP NNVKVAVCDI PPRGLKMAAT FIGNNTAIQE LFIRVSEQFS AMFRRKAFLH WYTG EGMDE MEFSEAEGNT NDLVSEYQQY QDATADVEEY EEAEASPEKE T UniProtKB: Tubulin beta-6 chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: Cryptophycin 52
| Macromolecule | Name: Cryptophycin 52 / type: ligand / ID: 4 / Number of copies: 8 / Formula: YGY |
|---|---|
| Molecular weight | Theoretical: 669.204 Da |
| Chemical component information | 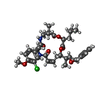 ChemComp-YGY: |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 8 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 0.1 M PIPES-KOH pH=7.0,1mM MgCL2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.88 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































