+Search query
-Structure paper
| Title | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1. |
|---|---|
| Journal, issue, pages | Nature, Vol. 629, Issue 8012, Page 710-716, Year 2024 |
| Publish date | May 1, 2024 |
 Authors Authors | Yeeun Son / Timothy C Kenny / Artem Khan / Kıvanç Birsoy / Richard K Hite /  |
| PubMed Abstract | Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. ...Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. Despite the essential roles of these lipids, the mechanisms that enable the cellular uptake of choline and ethanolamine remain unknown. Here we show that the protein encoded by FLVCR1, whose mutation leads to the neurodegenerative syndrome posterior column ataxia and retinitis pigmentosa, transports extracellular choline and ethanolamine into cells for phosphorylation by downstream kinases to initiate the Kennedy pathway. Structures of FLVCR1 in the presence of choline and ethanolamine reveal that both metabolites bind to a common binding site comprising aromatic and polar residues. Despite binding to a common site, FLVCR1 interacts in different ways with the larger quaternary amine of choline in and with the primary amine of ethanolamine. Structure-guided mutagenesis identified residues that are crucial for the transport of ethanolamine, but dispensable for choline transport, enabling functional separation of the entry points into the two branches of the Kennedy pathway. Altogether, these studies reveal how FLVCR1 is a high-affinity metabolite transporter that serves as the common origin for phospholipid biosynthesis by two branches of the Kennedy pathway. |
 External links External links |  Nature / Nature /  PubMed:38693265 / PubMed:38693265 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.42 - 3.02 Å |
| Structure data | EMDB-42107, PDB-8ubw: EMDB-42108, PDB-8ubx: EMDB-42109, PDB-8uby: EMDB-42110, PDB-8ubz: EMDB-42111, PDB-8uc0: |
| Chemicals |  ChemComp-Y01: 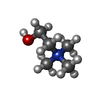 ChemComp-CHT:  ChemComp-HOH: 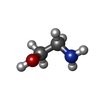 ChemComp-ETA: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / Choline / ethanolamine / membrane transport |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 homo sapiens (human)
homo sapiens (human)