+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8uby | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Choline-bound FLVCR1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
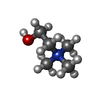
 Components Components | Heme transporter FLVCR1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Choline / ethanolamine / membrane transport | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationheme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth ...heme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth / Heme biosynthesis / heme biosynthetic process / embryonic digit morphogenesis / erythrocyte maturation / mitochondrial transport / blood vessel development / spleen development / erythrocyte differentiation / Iron uptake and transport / multicellular organism growth / in utero embryonic development / intracellular iron ion homeostasis / mitochondrial inner membrane / heme binding / mitochondrion / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.67 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Hite, R.K. / Son, Y. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support | 1items
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1. Authors: Yeeun Son / Timothy C Kenny / Artem Khan / Kıvanç Birsoy / Richard K Hite /  Abstract: Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. ...Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. Despite the essential roles of these lipids, the mechanisms that enable the cellular uptake of choline and ethanolamine remain unknown. Here we show that the protein encoded by FLVCR1, whose mutation leads to the neurodegenerative syndrome posterior column ataxia and retinitis pigmentosa, transports extracellular choline and ethanolamine into cells for phosphorylation by downstream kinases to initiate the Kennedy pathway. Structures of FLVCR1 in the presence of choline and ethanolamine reveal that both metabolites bind to a common binding site comprising aromatic and polar residues. Despite binding to a common site, FLVCR1 interacts in different ways with the larger quaternary amine of choline in and with the primary amine of ethanolamine. Structure-guided mutagenesis identified residues that are crucial for the transport of ethanolamine, but dispensable for choline transport, enabling functional separation of the entry points into the two branches of the Kennedy pathway. Altogether, these studies reveal how FLVCR1 is a high-affinity metabolite transporter that serves as the common origin for phospholipid biosynthesis by two branches of the Kennedy pathway. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8uby.cif.gz 8uby.cif.gz | 173.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8uby.ent.gz pdb8uby.ent.gz | 138.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8uby.json.gz 8uby.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ub/8uby https://data.pdbj.org/pub/pdb/validation_reports/ub/8uby ftp://data.pdbj.org/pub/pdb/validation_reports/ub/8uby ftp://data.pdbj.org/pub/pdb/validation_reports/ub/8uby | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  42109MC  8ubwC  8ubxC  8ubzC  8uc0C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 59899.352 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FLVCR1 / Production host: Homo sapiens (human) / Gene: FLVCR1 / Production host:  Homo sapiens (human) / References: UniProt: Q9Y5Y0 Homo sapiens (human) / References: UniProt: Q9Y5Y0 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Chemical | ChemComp-CHT / | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | Has protein modification | N | |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human FLVCR1 / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||
| Buffer solution | pH: 7.5 Details: 0.01 % LMNG, 0.001 % CHS, 20 mM HEPES (pHed with KOH, pH 7.5), 150 mM KCl | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Conc.: 6 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 297 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1500 nm / Nominal defocus min: 500 nm |
| Image recording | Electron dose: 53.98 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
| EM imaging optics | Energyfilter name: TFS Selectris X |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 6540900 | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.67 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 50496 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Protocol: BACKBONE TRACE / Space: REAL / Target criteria: FSC 0.5 | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj