+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Choline-bound FLVCR1 | |||||||||
 Map data Map data | Choline-bound FLVCR1 from 1 mM Ethanolamine data set | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Choline / ethanolamine / membrane transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationheme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth ...heme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth / Heme biosynthesis / heme biosynthetic process / embryonic digit morphogenesis / erythrocyte maturation / mitochondrial transport / blood vessel development / spleen development / erythrocyte differentiation / Iron uptake and transport / multicellular organism growth / in utero embryonic development / intracellular iron ion homeostasis / mitochondrial inner membrane / heme binding / mitochondrion / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
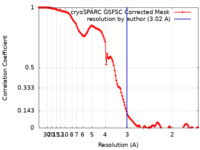
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Hite RK / Son Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1. Authors: Yeeun Son / Timothy C Kenny / Artem Khan / Kıvanç Birsoy / Richard K Hite /  Abstract: Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. ...Phosphatidylcholine and phosphatidylethanolamine, the two most abundant phospholipids in mammalian cells, are synthesized de novo by the Kennedy pathway from choline and ethanolamine, respectively. Despite the essential roles of these lipids, the mechanisms that enable the cellular uptake of choline and ethanolamine remain unknown. Here we show that the protein encoded by FLVCR1, whose mutation leads to the neurodegenerative syndrome posterior column ataxia and retinitis pigmentosa, transports extracellular choline and ethanolamine into cells for phosphorylation by downstream kinases to initiate the Kennedy pathway. Structures of FLVCR1 in the presence of choline and ethanolamine reveal that both metabolites bind to a common binding site comprising aromatic and polar residues. Despite binding to a common site, FLVCR1 interacts in different ways with the larger quaternary amine of choline in and with the primary amine of ethanolamine. Structure-guided mutagenesis identified residues that are crucial for the transport of ethanolamine, but dispensable for choline transport, enabling functional separation of the entry points into the two branches of the Kennedy pathway. Altogether, these studies reveal how FLVCR1 is a high-affinity metabolite transporter that serves as the common origin for phospholipid biosynthesis by two branches of the Kennedy pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42110.map.gz emd_42110.map.gz | 108.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42110-v30.xml emd-42110-v30.xml emd-42110.xml emd-42110.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42110_fsc.xml emd_42110_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42110.png emd_42110.png | 117.2 KB | ||
| Filedesc metadata |  emd-42110.cif.gz emd-42110.cif.gz | 7 KB | ||
| Others |  emd_42110_additional_1.map.gz emd_42110_additional_1.map.gz emd_42110_half_map_1.map.gz emd_42110_half_map_1.map.gz emd_42110_half_map_2.map.gz emd_42110_half_map_2.map.gz | 4.3 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42110 http://ftp.pdbj.org/pub/emdb/structures/EMD-42110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42110 | HTTPS FTP |
-Related structure data
| Related structure data |  8ubzMC  8ubwC  8ubxC  8ubyC  8uc0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42110.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42110.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Choline-bound FLVCR1 from 1 mM Ethanolamine data set | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Density modified map
| File | emd_42110_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_42110_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_42110_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human FLVCR1
| Entire | Name: Human FLVCR1 |
|---|---|
| Components |
|
-Supramolecule #1: Human FLVCR1
| Supramolecule | Name: Human FLVCR1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Heme transporter FLVCR1
| Macromolecule | Name: Heme transporter FLVCR1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.899352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARPDDEEGA AVAPGHPLAK GYLPLPRGAP VGKESVELQN GPKAGTFPVN GAPRDSLAAA SGVLGGPQTP LAPEEETQAR LLPAGAGAE TPGAESSPLP LTALSPRRFV VLLIFSLYSL VNAFQWIQYS IISNVFEGFY GVTLLHIDWL SMVYMLAYVP L IFPATWLL ...String: MARPDDEEGA AVAPGHPLAK GYLPLPRGAP VGKESVELQN GPKAGTFPVN GAPRDSLAAA SGVLGGPQTP LAPEEETQAR LLPAGAGAE TPGAESSPLP LTALSPRRFV VLLIFSLYSL VNAFQWIQYS IISNVFEGFY GVTLLHIDWL SMVYMLAYVP L IFPATWLL DTRGLRLTAL LGSGLNCLGA WIKCGSVQQH LFWVTMLGQC LCSVAQVFIL GLPSRIASVW FGPKEVSTAC AT AVLGNQL GTAVGFLLPP VLVPNTQNDT NLLACNISTM FYGTSAVATL LFILTAIAFK EKPRYPPSQA QAALQDSPPE EYS YKKSIR NLFKNIPFVL LLITYGIMTG AFYSVSTLLN QMILTYYEGE EVNAGRIGLT LVVAGMVGSI LCGLWLDYTK TYKQ TTLIV YILSFIGMVI FTFTLDLRYI IIVFVTGGVL GFFMTGYLPL GFEFAVEITY PESEGTSSGL LNASAQIFGI LFTLA QGKL TSDYGPKAGN IFLCVWMFIG IILTALIKSD LRRHNINIGI TNVDVKAIPA DSPTDQEPKT VMLSKQSESA I UniProtKB: Choline/ethanolamine transporter FLVCR1 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 3 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: CHOLINE ION
| Macromolecule | Name: CHOLINE ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: CHT |
|---|---|
| Molecular weight | Theoretical: 104.171 Da |
| Chemical component information | 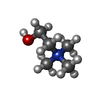 ChemComp-CHT: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 26 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.0 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 0.01 % LMNG, 0.001 % CHS, 20 mM HEPES (pHed with KOH, pH 7.5), 150 mM KCl, 1 mM ethanol amine | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: FSC 0.5 |
|---|---|
| Output model |  PDB-8ubz: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)