+Search query
-Structure paper
| Title | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 43, Issue 11, Page 114895, Year 2024 |
| Publish date | Nov 26, 2024 |
 Authors Authors | Fang Ye / Xin Pan / Zhiyi Zhang / Xufu Xiang / Xinyu Li / Binghao Zhang / Peiruo Ning / Aijun Liu / Qinggong Wang / Kaizheng Gong / Jiancheng Li / Lizhe Zhu / Chungen Qian / Geng Chen / Yang Du /  |
| PubMed Abstract | Hydroxycarboxylic acid receptor 3 (HCAR3), a class A G-protein-coupled receptor, is an important cellular energy metabolism sensor with a key role in the regulation of lipolysis in humans. HCAR3 is ...Hydroxycarboxylic acid receptor 3 (HCAR3), a class A G-protein-coupled receptor, is an important cellular energy metabolism sensor with a key role in the regulation of lipolysis in humans. HCAR3 is deeply involved in many physiological processes and serves as a valuable target for the treatment of metabolic diseases, tumors, and immune diseases. Here, we report four cryoelectron microscopy (cryo-EM) structures of human HCAR3-Gi1 complexes with or without agonists: the endogenous ligand 3-hydroxyoctanoic acid, the drug niacin, the highly subtype-specific agonist compound 5c (4-(n-propyl)amino-3-nitrobenzoic acid), and the apo form. Together with mutagenesis and functional analyses, we revealed the recognition mechanisms of HCAR3 for different agonists. In addition, the key residues that determine the ligand selectivity between HCAR2 and HCAR3 were also illuminated. Overall, these findings provide a structural basis for the ligand recognition, activation, and selectivity and G-protein coupling mechanisms of HCAR3, which contribute to the design of HCAR3-targeting drugs with high efficacy and selectivity. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:39427321 PubMed:39427321 |
| Methods | EM (single particle) |
| Resolution | 2.73 - 3.4 Å |
| Structure data | EMDB-36186, PDB-8jef: EMDB-36189, PDB-8jei: EMDB-61501, PDB-9jic: EMDB-61502, PDB-9jid: |
| Chemicals | 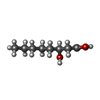 ChemComp-3HO: 
ChemComp-CW3:  ChemComp-NIO: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / complex / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)