+Search query
-Structure paper
| Title | Structural basis and dynamics of Chikungunya alphavirus RNA capping by nsP1 capping pores. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 120, Issue 12, Page e2213934120, Year 2023 |
| Publish date | Mar 21, 2023 |
 Authors Authors | Rhian Jones / Michael Hons / Nadia Rabah / Noelia Zamarreño / Rocío Arranz / Juan Reguera /   |
| PubMed Abstract | Alphaviruses are emerging positive-stranded RNA viruses which replicate and transcribe their genomes in membranous organelles formed in the cell cytoplasm. The nonstructural protein 1 (nsP1) is ...Alphaviruses are emerging positive-stranded RNA viruses which replicate and transcribe their genomes in membranous organelles formed in the cell cytoplasm. The nonstructural protein 1 (nsP1) is responsible for viral RNA capping and gates the replication organelles by assembling into monotopic membrane-associated dodecameric pores. The capping pathway is unique to Alphaviruses; beginning with the N methylation of a guanosine triphosphate (GTP) molecule, followed by the covalent linkage of an mGMP group to a conserved histidine in nsP1 and the transfer of this cap structure to a diphosphate RNA. Here, we provide structural snapshots of different stages of the reaction pathway showing how nsP1 pores recognize the substrates of the methyl-transfer reaction, GTP and S-adenosyl methionine (SAM), how the enzyme reaches a metastable postmethylation state with SAH and mGTP in the active site, and the subsequent covalent transfer of mGMP to nsP1 triggered by the presence of RNA and postdecapping reaction conformational changes inducing the opening of the pore. In addition, we biochemically characterize the capping reaction, demonstrating specificity for the RNA substrate and the reversibility of the cap transfer resulting in decapping activity and the release of reaction intermediates. Our data identify the molecular determinants allowing each pathway transition, providing an explanation for the need for the SAM methyl donor all along the pathway and clues about the conformational rearrangements associated to the enzymatic activity of nsP1. Together, our results set ground for the structural and functional understanding of alphavirus RNA-capping and the design of antivirals. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:36913573 / PubMed:36913573 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.48 - 3.2 Å |
| Structure data | EMDB-15553, PDB-8aov: EMDB-15554, PDB-8aow: EMDB-15555, PDB-8aox: EMDB-15578, PDB-8apx: EMDB-15704, PDB-8axv: |
| Chemicals |  ChemComp-ZN:  ChemComp-GTP:  ChemComp-HOH:  ChemComp-SAH: 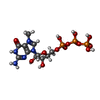 ChemComp-MGP:  ChemComp-MG:  ChemComp-SAM: 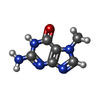 ChemComp-MY6: |
| Source |
|
 Keywords Keywords | VIRAL PROTEIN / Alphavirus Replication complex capping pores membrane pore Methyltransferase Guanylyl transferase / Alphavirus Replication complex Capping pores Membrane pore Methyltransferase gunayltransferase VIRAL PROTEIN / Alphavirus Replication complex / capping pores / membrane pore / Methyltransferase / Guanylyl transferase / VIRAL PROT |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 Baculovirus expression vector pFastBac1-HM
Baculovirus expression vector pFastBac1-HM