+Search query
-Structure paper
| Title | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 4405, Year 2023 |
| Publish date | Jul 21, 2023 |
 Authors Authors | Zhennan Zhao / Yufeng Xie / Bin Bai / Chunliang Luo / Jingya Zhou / Weiwei Li / Yumin Meng / Linjie Li / Dedong Li / Xiaomei Li / Xiaoxiong Li / Xiaoyun Wang / Junqing Sun / Zepeng Xu / Yeping Sun / Wei Zhang / Zheng Fan / Xin Zhao / Linhuan Wu / Juncai Ma / Odel Y Li / Guijun Shang / Yan Chai / Kefang Liu / Peiyi Wang / George F Gao / Jianxun Qi /  |
| PubMed Abstract | Multiple SARS-CoV-2 Omicron sub-variants, such as BA.2, BA.2.12.1, BA.4, and BA.5, emerge one after another. BA.5 has become the dominant strain worldwide. Additionally, BA.2.75 is significantly ...Multiple SARS-CoV-2 Omicron sub-variants, such as BA.2, BA.2.12.1, BA.4, and BA.5, emerge one after another. BA.5 has become the dominant strain worldwide. Additionally, BA.2.75 is significantly increasing in some countries. Exploring their receptor binding and interspecies transmission risk is urgently needed. Herein, we examine the binding capacities of human and other 28 animal ACE2 orthologs covering nine orders towards S proteins of these sub-variants. The binding affinities between hACE2 and these sub-variants remain in the range as that of previous variants of concerns (VOCs) or interests (VOIs). Notably, R493Q reverse mutation enhances the bindings towards ACE2s from humans and many animals closely related to human life, suggesting an increased risk of cross-species transmission. Structures of S/hACE2 or RBD/hACE2 complexes for these sub-variants and BA.2 S binding to ACE2 of mouse, rat or golden hamster are determined to reveal the molecular basis for receptor binding and broader interspecies recognition. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37479708 / PubMed:37479708 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.58 - 3.29 Å |
| Structure data | EMDB-33841, PDB-7yhw: EMDB-33870, PDB-7yj3: EMDB-34120, PDB-7yv8: EMDB-34138, PDB-7yvu: EMDB-34217, PDB-8gry: EMDB-34409, PDB-8h06:  EMDB-34494: Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with human ACE2 (three-RBD-up conformation)  EMDB-34498: Cryo-EM map of SARS-CoV-2 Omicron BA.2.12.1 spike trimer in complex with human ACE2 (three-RBD-up conformation)  EMDB-34499: Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2  EMDB-34506: Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with rat ACE2  EMDB-34509: Cryo-EM map of SARS-CoV-2 Omicron BA.4/5 (N658S) spike trimer in complex with human ACE2 (three-RBD-up conformation) 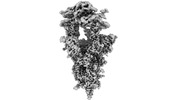 EMDB-34510: Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with golden hamster ACE2  PDB-8h5c: |
| Chemicals |  ChemComp-NAG:  ChemComp-ZN: |
| Source |
|
 Keywords Keywords | HYDROLASE/VIRAL PROTEIN / SARS-CoV-2 / Omicron BA.2.12.1 / spike protein / VIRAL PROTEIN / HYDROLASE-VIRAL PROTEIN complex / Omicron BA.2 / Omicron BA.4/5 / Omicron BA.2.75 / hACE2 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



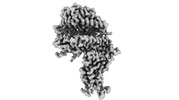

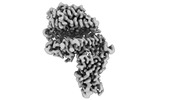

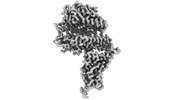








 homo sapiens (human)
homo sapiens (human) mesocricetus auratus (golden hamster)
mesocricetus auratus (golden hamster)