+Search query
-Structure paper
| Title | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca2.2. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 37, Issue 5, Page 109931, Year 2021 |
| Publish date | Nov 2, 2021 |
 Authors Authors | Yanli Dong / Yiwei Gao / Shuai Xu / Yuhang Wang / Zhuoya Yu / Yue Li / Bin Li / Tian Yuan / Bei Yang / Xuejun Cai Zhang / Daohua Jiang / Zhuo Huang / Yan Zhao /  |
| PubMed Abstract | N-type voltage-gated calcium (Ca) channels mediate Ca influx at presynaptic terminals in response to action potentials and play vital roles in synaptogenesis, release of neurotransmitters, and ...N-type voltage-gated calcium (Ca) channels mediate Ca influx at presynaptic terminals in response to action potentials and play vital roles in synaptogenesis, release of neurotransmitters, and nociceptive transmission. Here, we elucidate a cryo-electron microscopy (cryo-EM) structure of the human Ca2.2 complex in apo, ziconotide-bound, and two Ca2.2-specific pore blockers-bound states. The second voltage-sensing domain (VSD) is captured in a resting-state conformation, trapped by a phosphatidylinositol 4,5-bisphosphate (PIP) molecule, which is distinct from the other three VSDs of Ca2.2, as well as activated VSDs observed in previous structures of Ca channels. This structure reveals the molecular basis for the unique inactivation process of Ca2.2 channels, in which the intracellular gate formed by S6 helices is closed and a W-helix from the domain II-III linker stabilizes closed-state inactivation. The structures of this inactivated, drug-bound complex lay a solid foundation for developing new state-dependent blockers for treatment of chronic pain. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:34731621 PubMed:34731621 |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.3 Å |
| Structure data | EMDB-31958, PDB-7vfs: EMDB-31959, PDB-7vfu: EMDB-31960, PDB-7vfv: EMDB-31961, PDB-7vfw: |
| Chemicals |  ChemComp-R16:  ChemComp-Y01:  ChemComp-PT5:  ChemComp-CA:  ChemComp-NAG: 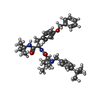 ChemComp-6IX: 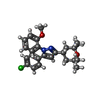 ChemComp-6I7: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Voltage gated calcium channel / N-type / complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human) conus magus (magus cone)
conus magus (magus cone)