+Search query
-Structure paper
| Title | Structural insights into ligand recognition, activation, and signaling of the α adrenergic receptor. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 8, Issue 9, Page eabj5347, Year 2022 |
| Publish date | Mar 4, 2022 |
 Authors Authors | Jun Xu / Sheng Cao / Harald Hübner / Dorothée Weikert / Geng Chen / Qiuyuan Lu / Daopeng Yuan / Peter Gmeiner / Zheng Liu / Yang Du /   |
| PubMed Abstract | The α adrenergic receptor (αAR) is a G protein (heterotrimeric guanine nucleotide-binding protein)-coupled receptor that mediates important physiological functions in response to the endogenous ...The α adrenergic receptor (αAR) is a G protein (heterotrimeric guanine nucleotide-binding protein)-coupled receptor that mediates important physiological functions in response to the endogenous neurotransmitters norepinephrine and epinephrine, as well as numerous chemically distinct drugs. However, the molecular mechanisms of drug actions remain poorly understood. Here, we report the cryo-electron microscopy structures of the human αAR-GoA complex bound to norepinephrine and three imidazoline derivatives (brimonidine, dexmedetomidine, and oxymetazoline). Together with mutagenesis and functional data, these structures provide important insights into the molecular basis of ligand recognition, activation, and signaling at the αAR. Further structural analyses uncover different molecular determinants between αAR and βARs for recognition of norepinephrine and key regions that determine the G protein coupling selectivity. Overall, our studies provide a framework for understanding the signal transduction of the adrenergic system at the atomic level, which will facilitate rational structure-based discovery of safer and more effective medications for αAR. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:35245122 / PubMed:35245122 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.0 - 3.6 Å |
| Structure data | EMDB-31147, PDB-7ej0: EMDB-31156, PDB-7ej8: EMDB-31157, PDB-7eja: EMDB-31162, PDB-7ejk: |
| Chemicals |  ChemComp-E5E: 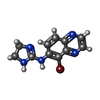 ChemComp-J59: 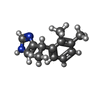 ChemComp-CZX: 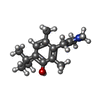 ChemComp-J5C: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / alpha2A-adrenergic receptor / signaling complex / cryo-EM |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)
