+Search query
-Structure paper
| Title | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. |
|---|---|
| Journal, issue, pages | Science, Vol. 368, Issue 6496, Page 1211-1219, Year 2020 |
| Publish date | Jun 12, 2020 |
 Authors Authors | Lu Zhang / Yao Zhao / Yan Gao / Lijie Wu / Ruogu Gao / Qi Zhang / Yinan Wang / Chengyao Wu / Fangyu Wu / Sudagar S Gurcha / Natacha Veerapen / Sarah M Batt / Wei Zhao / Ling Qin / Xiuna Yang / Manfu Wang / Yan Zhu / Bing Zhang / Lijun Bi / Xian'en Zhang / Haitao Yang / Luke W Guddat / Wenqing Xu / Quan Wang / Jun Li / Gurdyal S Besra / Zihe Rao /    |
| PubMed Abstract | The arabinosyltransferases EmbA, EmbB, and EmbC are involved in cell wall synthesis and are recognized as targets for the anti-tuberculosis drug ethambutol. In this study, we determined cryo- ...The arabinosyltransferases EmbA, EmbB, and EmbC are involved in cell wall synthesis and are recognized as targets for the anti-tuberculosis drug ethambutol. In this study, we determined cryo-electron microscopy and x-ray crystal structures of mycobacterial EmbA-EmbB and EmbC-EmbC complexes in the presence of their glycosyl donor and acceptor substrates and with ethambutol. These structures show how the donor and acceptor substrates bind in the active site and how ethambutol inhibits arabinosyltransferases by binding to the same site as both substrates in EmbB and EmbC. Most drug-resistant mutations are located near the ethambutol binding site. Collectively, our work provides a structural basis for understanding the biochemical function and inhibition of arabinosyltransferases and the development of new anti-tuberculosis agents. |
 External links External links |  Science / Science /  PubMed:32327601 PubMed:32327601 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.81 - 3.3 Å |
| Structure data | EMDB-30216, PDB-7bvc: EMDB-30217, PDB-7bve: EMDB-30218, PDB-7bvf: EMDB-30219, PDB-7bvg:  PDB-7bvh: |
| Chemicals |  ChemComp-CDL: 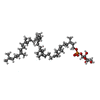 ChemComp-F8L:  ChemComp-CA:  ChemComp-PNS:  ChemComp-PO4: 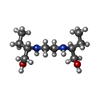 ChemComp-95E:  ChemComp-PN7:  ChemComp-DSL: 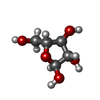 ChemComp-BXY: |
| Source |
|
 Keywords Keywords | TRANSFERASE / Mycobacterium smegmatis / cell wall synthesis / drug target / ethambutol / arabinosyltransferase / EmbA / EmbB / EmbC / acyl carrier protein / arabinogalactan / lipoarabinomannan / drug resistance / Mycobacterium tuberculosis |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 mycolicibacterium smegmatis mc2 155 (bacteria)
mycolicibacterium smegmatis mc2 155 (bacteria)