+Search query
-Structure paper
| Title | Structural Analysis and Development of Notum Fragment Screening Hits. |
|---|---|
| Journal, issue, pages | Acs Chem Neurosci, Vol. 13, Page 2060-2077, Year 2022 |
| Publish date | Dec 2, 2020 (structure data deposition date) |
 Authors Authors | Zhao, Y. / Mahy, W. / Willis, N.J. / Woodward, H.L. / Steadman, D. / Bayle, E.D. / Atkinson, B.N. / Sipthorp, J. / Vecchia, L. / Ruza, R.R. ...Zhao, Y. / Mahy, W. / Willis, N.J. / Woodward, H.L. / Steadman, D. / Bayle, E.D. / Atkinson, B.N. / Sipthorp, J. / Vecchia, L. / Ruza, R.R. / Harlos, K. / Jeganathan, F. / Constantinou, S. / Costa, A. / Kjaer, S. / Bictash, M. / Salinas, P.C. / Whiting, P. / Vincent, J.P. / Fish, P.V. / Jones, E.Y. |
 External links External links |  Acs Chem Neurosci / Acs Chem Neurosci /  PubMed:35731924 PubMed:35731924 |
| Methods | X-ray diffraction |
| Resolution | 1.24 - 1.94 Å |
| Structure data |  PDB-7b4x:  PDB-7b84:  PDB-7b86:  PDB-7b87:  PDB-7b89:  PDB-7b8c:  PDB-7b8d:  PDB-7b8f:  PDB-7b8g:  PDB-7b8j:  PDB-7b8k:  PDB-7b8l:  PDB-7b8m:  PDB-7b8n:  PDB-7b8o:  PDB-7b8u:  PDB-7b8x:  PDB-7b8y:  PDB-7b8z:  PDB-7b98:  PDB-7b99:  PDB-7b9d:  PDB-7b9i:  PDB-7b9n:  PDB-7b9u:  PDB-7ba1:  PDB-7bac:  PDB-7bap:  PDB-7bc8:  PDB-7bc9:  PDB-7bcc:  PDB-7bcd:  PDB-7bce:  PDB-7bcf:  PDB-7bch:  PDB-7bci:  PDB-7bck:  PDB-7bcl:  PDB-7bd2:  PDB-7bd3:  PDB-7bd4:  PDB-7bd5:  PDB-7bd6:  PDB-7bd8:  PDB-7bd9:  PDB-7bda:  PDB-7bdb:  PDB-7bdc:  PDB-7bdd:  PDB-7bdf:  PDB-7bdg:  PDB-7bdh: |
| Chemicals |  ChemComp-SO4:  ChemComp-DMS:  ChemComp-EDO:  ChemComp-SWT:  ChemComp-HOH:  ChemComp-NAG:  ChemComp-JFS:  ChemComp-AW7: 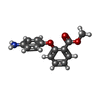 ChemComp-JFV: 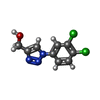 ChemComp-T1Z: 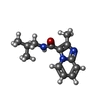 ChemComp-GRV: 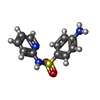 ChemComp-SFY:  ChemComp-RYM: 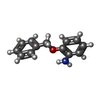 ChemComp-MJW: 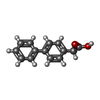 ChemComp-BP4:  ChemComp-AXV:  ChemComp-AY4: 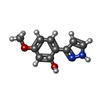 ChemComp-GQM:  ChemComp-GOL:  ChemComp-T2E:  ChemComp-T1W:  ChemComp-RUY:  ChemComp-U3K:  ChemComp-T2H: 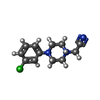 ChemComp-A6Z: 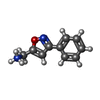 ChemComp-EJW:  ChemComp-B1A: 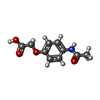 ChemComp-T3N: 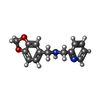 ChemComp-T3T:  ChemComp-N9J:  ChemComp-O3D:  ChemComp-S1V: 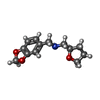 ChemComp-LO5: 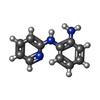 ChemComp-S4V: 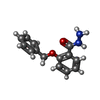 ChemComp-TAH:  ChemComp-UR4: 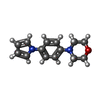 ChemComp-TB5: 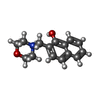 ChemComp-T9Z:  ChemComp-LDV:  ChemComp-T9W:  ChemComp-T9Q:  ChemComp-GVV:  ChemComp-T9B:  ChemComp-T9K:  ChemComp-NUM:  ChemComp-TE5:  ChemComp-JHJ:  ChemComp-YI6:  ChemComp-JH7:  ChemComp-QL5:  ChemComp-GW1:  ChemComp-O1J:  ChemComp-NW7:  ChemComp-WNA:  ChemComp-TE8:  ChemComp-TEH:  ChemComp-TEK:  ChemComp-TEZ: |
| Source |
|
 Keywords Keywords | HYDROLASE / Notum Inhibitor complex / Notum Fragment / Notum Inhibitor / Notum Inhibitor Wnt / Notum Inhibitor Fragment Screen |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)