+Search query
-Structure paper
| Title | Structure and superorganization of acetylcholine receptor-rapsyn complexes. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 110, Issue 26, Page 10622-10627, Year 2013 |
| Publish date | Jun 25, 2013 |
 Authors Authors | Benoît Zuber / Nigel Unwin /  |
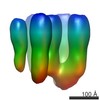
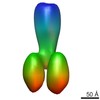
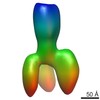
| PubMed Abstract | The scaffolding protein at the neuromuscular junction, rapsyn, enables clustering of nicotinic acetylcholine receptors in high concentration and is critical for muscle function. Patients with ...The scaffolding protein at the neuromuscular junction, rapsyn, enables clustering of nicotinic acetylcholine receptors in high concentration and is critical for muscle function. Patients with insufficient receptor clustering suffer from muscle weakness. However, the detailed organization of the receptor-rapsyn network is poorly understood: it is unclear whether rapsyn first forms a wide meshwork to which receptors can subsequently dock or whether it only forms short bridges linking receptors together to make a large cluster. Furthermore, the number of rapsyn-binding sites per receptor (a heteropentamer) has been controversial. Here, we show by cryoelectron tomography and subtomogram averaging of Torpedo postsynaptic membrane that receptors are connected by up to three rapsyn bridges, the minimum number required to form a 2D network. Half of the receptors belong to rapsyn-connected groups comprising between two and fourteen receptors. Our results provide a structural basis for explaining the stability and low diffusion of receptors within clusters. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:23754381 / PubMed:23754381 /  PubMed Central PubMed Central |
| Methods | EM (subtomogram averaging) / EM (tomography) |
| Resolution | 40 - 50 Å |
| Structure data | EMDB-2376, PDB-4bog: EMDB-2377: The structure and super-organization of acetylcholine receptor-rapsyn complexes; class A EMDB-2378: The structure and super-organization of acetylcholine receptor-rapsyn complexes; class B EMDB-2381: The structure and super-organization of acetylcholine receptor-rapsyn complexes; class C |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / CLUSTERING / SYNAPSE / NEUROMUSCULAR JUNCTION / NICOTINIC / RAPSYN / 43K / ELECTRIC ORGAN / NEUROTRANSMITTER RECEPTOR / RECEPTOR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers