+Search query
-Structure paper
| Title | Systematic metabolite screening identifies functional regulators of the adenosine A2A receptor. |
|---|---|
| Journal, issue, pages | Commun Chem, Vol. 9, Issue 1, Page 19, Year 2025 |
| Publish date | Dec 9, 2025 |
 Authors Authors | Prashant Rao / Manoj Rathinaswamy / Michelle Chan / Andrea Guzmán Paredes / Chirag Patel / Bernd J Wranik / Jonathan Powell / Dan Eaton / Jared Rutter / Kevin G Hicks / Amirhossein Mafi / Qi Hao /  |
| PubMed Abstract | The adenosine A2A receptor (A2AR) is a Class A G protein-coupled receptor (GPCR) that regulates inflammation, glucose metabolism, and energy homeostasis in metabolically active tissues. While the ...The adenosine A2A receptor (A2AR) is a Class A G protein-coupled receptor (GPCR) that regulates inflammation, glucose metabolism, and energy homeostasis in metabolically active tissues. While the effects of small-molecule ligands and protein interactions with A2AR have been extensively studied, the regulatory influence of endogenous metabolites remains unexplored. To address this gap, we employed the Mass spectrometry Integrated with equilibrium Dialysis for the discovery of Allostery Systematically (MIDAS) platform to screen a library of human metabolites for interactions with A2AR. This approach identified 180 metabolites that interact with A2AR, including allosteric and orthosteric modulators. We characterized the mechanisms of three metabolites previously unreported to interact with A2AR: prostaglandin D2, an allosteric antagonist that fully inhibits receptor signaling, and two orthosteric agonists, S-adenosyl-L-homocysteine and 2'-deoxyadenosine, that fully activate A2AR. Overall, these findings highlight the potential of the MIDAS platform to uncover previously unrecognized metabolite-GPCR interactions for research and therapeutic applications. |
 External links External links |  Commun Chem / Commun Chem /  PubMed:41366530 / PubMed:41366530 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.15 - 3.34 Å |
| Structure data | EMDB-71130: A2AR-BRIL + ZM241385 + PGD2 EMDB-71131, PDB-9p1t: |
| Chemicals |  ChemComp-LMN: 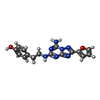 ChemComp-ZMA: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human)