+Search query
-Structure paper
| Title | Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. |
|---|---|
| Journal, issue, pages | Protein Cell, Vol. 9, Issue 6, Page 553-567, Year 2018 |
| Publish date | Mar 28, 2018 |
 Authors Authors | Jing-Xiang Wu / Dian Ding / Mengmeng Wang / Yunlu Kang / Xin Zeng / Lei Chen /  |
| PubMed Abstract | ATP-sensitive potassium channels (K) are energy sensors on the plasma membrane. By sensing the intracellular ADP/ATP ratio of β-cells, pancreatic K channels control insulin release and regulate ...ATP-sensitive potassium channels (K) are energy sensors on the plasma membrane. By sensing the intracellular ADP/ATP ratio of β-cells, pancreatic K channels control insulin release and regulate metabolism at the whole body level. They are implicated in many metabolic disorders and diseases and are therefore important drug targets. Here, we present three structures of pancreatic K channels solved by cryo-electron microscopy (cryo-EM), at resolutions ranging from 4.1 to 4.5 Å. These structures depict the binding site of the antidiabetic drug glibenclamide, indicate how Kir6.2 (inward-rectifying potassium channel 6.2) N-terminus participates in the coupling between the peripheral SUR1 (sulfonylurea receptor 1) subunit and the central Kir6.2 channel, reveal the binding mode of activating nucleotides, and suggest the mechanism of how Mg-ADP binding on nucleotide binding domains (NBDs) drives a conformational change of the SUR1 subunit. |
 External links External links |  Protein Cell / Protein Cell /  PubMed:29594720 / PubMed:29594720 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 4.11 - 6.1 Å |
| Structure data | EMDB-6831, PDB-5yke: EMDB-6832, PDB-5ykf: EMDB-6833, PDB-5ykg: EMDB-6847, PDB-5yw7: EMDB-6848, PDB-5yw8: EMDB-6849, PDB-5yw9: EMDB-6850, PDB-5ywa: EMDB-6851, PDB-5ywb: |
| Chemicals | 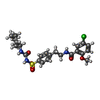 ChemComp-GBM:  ChemComp-AGS:  ChemComp-ADP:  ChemComp-MG: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / KATP / channel / glibenclamide / sulfonylurea |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
























