+Search query
-Structure paper
| Title | Distinct oligomeric assemblies of STING induced by non-nucleotide agonists. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 16, Issue 1, Page 3440, Year 2025 |
| Publish date | Apr 11, 2025 |
 Authors Authors | Anant Gharpure / Ariana Sulpizio / Johannes R Loeffler / Monica L Fernández-Quintero / Andy S Tran / Luke L Lairson / Andrew B Ward /  |
| PubMed Abstract | STING plays essential roles coordinating innate immune responses to processes that range from pathogenic infection to genomic instability. Its adaptor function is activated by cyclic dinucleotide ...STING plays essential roles coordinating innate immune responses to processes that range from pathogenic infection to genomic instability. Its adaptor function is activated by cyclic dinucleotide (CDN) secondary messengers originating from self (2'3'-cGAMP) or bacterial sources (3'3'-CDNs). Different classes of CDNs possess distinct binding modes, stabilizing STING's ligand-binding domain (LBD) in either a closed or open conformation. The closed conformation, induced by the endogenous ligand 2'3'-cGAMP, has been extensively studied using cryo-EM. However, significant questions remain regarding the structural basis of STING activation by open conformation-inducing ligands. Using cryo-EM, we investigate potential differences in conformational changes and oligomeric assemblies of STING for closed and open conformation-inducing synthetic agonists. While we observe a characteristic 180° rotation for both classes, the open-LBD inducing agonist diABZI-3 uniquely induces a quaternary structure reminiscent but distinct from the reported autoinhibited state of apo-STING. Additionally, we observe slower rates of activation for this ligand class in functional assays, which collectively suggests the existence of a potential additional regulatory mechanism for open conformation-inducing ligands that involves head-to-head interactions and restriction of curved oligomer formation. These observations have potential implications in the selection of an optimal class of STING agonist in the context of a defined therapeutic application. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:40216780 / PubMed:40216780 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.09 - 3.67 Å |
| Structure data | EMDB-45897, PDB-9ct3: EMDB-45898, PDB-9ct4: EMDB-45899, PDB-9ct5: EMDB-45900, PDB-9ct6: |
| Chemicals | 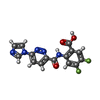 ChemComp-V67: 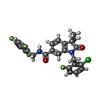 ChemComp-9IM:  PDB-1az0: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / Innate immunity / membrane protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)