+Search query
-Structure paper
| Title | Calcineurin-fusion facilitates cryo-EM structure determination of a Family A GPCR. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 121, Issue 48, Page e2414544121, Year 2024 |
| Publish date | Nov 26, 2024 |
 Authors Authors | Jun Xu / Geng Chen / Haoqing Wang / Sheng Cao / Jie Heng / Xavier Deupi / Yang Du / Brian K Kobilka /    |
| PubMed Abstract | Advances in singe-particle cryo-electron microscopy (cryo-EM) have made it possible to solve the structures of numerous Family A and Family B G protein-coupled receptors (GPCRs) in complex with G ...Advances in singe-particle cryo-electron microscopy (cryo-EM) have made it possible to solve the structures of numerous Family A and Family B G protein-coupled receptors (GPCRs) in complex with G proteins and arrestins, as well as several Family C GPCRs. Determination of these structures has been facilitated by the presence of large extramembrane components (such as G protein, arrestin, or Venus flytrap domains) in these complexes that aid in particle alignment during the processing of the cryo-EM data. In contrast, determination of the inactive state structure of Family A GPCRs is more challenging due to the relatively small size of the seven transmembrane domain (7TM) and to the surrounding detergent micelle that, in the absence of other features, make particle alignment impossible. Here, we describe an alternative protein engineering strategy where the heterodimeric protein calcineurin is fused to a GPCR by three points of attachment, the cytoplasmic ends of TM5, TM6, and TM7. This three-point attachment provides a more rigid link with the GPCR transmembrane domain that facilitates particle alignment during data processing, allowing us to determine the structures of the β adrenergic receptor (βAR) in the apo, antagonist-bound, and agonist-bound states. We expect that this fusion strategy may have broad application in cryo-EM structural determination of other Family A GPCRs. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:39565314 / PubMed:39565314 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.49 - 3.95 Å |
| Structure data | EMDB-45602, PDB-9chu: EMDB-45603, PDB-9chv: EMDB-45604, PDB-9chx: |
| Chemicals |  ChemComp-E5E: 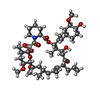 ChemComp-FK5: 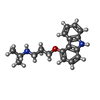 ChemComp-CAU: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN/HYDROLASE/ISOMERASE / GPCR / beta2AR / calcineurin / inactive state / MEMBRANE PROTEIN / MEMBRANE PROTEIN-HYDROLASE-ISOMERASE complex / cryo-EM / calcineurin fusion |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)
