+Search query
-Structure paper
| Title | Cryo-EM structure of the serotonin 5-HT receptor coupled to heterotrimeric G. |
|---|---|
| Journal, issue, pages | Nature, Vol. 558, Issue 7711, Page 620-623, Year 2018 |
| Publish date | Jun 20, 2018 |
 Authors Authors | Javier García-Nafría / Rony Nehmé / Patricia C Edwards / Christopher G Tate /  |
| PubMed Abstract | G-protein-coupled receptors (GPCRs) form the largest family of receptors encoded by the human genome (around 800 genes). They transduce signals by coupling to a small number of heterotrimeric G ...G-protein-coupled receptors (GPCRs) form the largest family of receptors encoded by the human genome (around 800 genes). They transduce signals by coupling to a small number of heterotrimeric G proteins (16 genes encoding different α-subunits). Each human cell contains several GPCRs and G proteins. The structural determinants of coupling of G to four different GPCRs have been elucidated, but the molecular details of how the other G-protein classes couple to GPCRs are unknown. Here we present the cryo-electron microscopy structure of the serotonin 5-HT receptor (5-HTR) bound to the agonist donitriptan and coupled to an engineered G heterotrimer. In this complex, 5-HTR is in an active state; the intracellular domain of the receptor is in a similar conformation to that observed for the β-adrenoceptor (βAR) or the adenosine A receptor (AR) in complex with G. In contrast to the complexes with G, the gap between the receptor and the Gβ-subunit in the G-5-HTR complex precludes molecular contacts, and the interface between the Gα-subunit of G and the receptor is considerably smaller. These differences are likely to be caused by the differences in the interactions with the C terminus of the G α-subunit. The molecular variations between the interfaces of G and G in complex with GPCRs may contribute substantially to both the specificity of coupling and the kinetics of signalling. |
 External links External links |  Nature / Nature /  PubMed:29925951 / PubMed:29925951 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.78 Å |
| Structure data | |
| Chemicals | 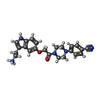 ChemComp-EP5: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / G-protein coupled receptor / 5-HT1B / Mini-Go / serotonin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)