+Search query
-Structure paper
| Title | Structural insights into vesicular monoamine storage and drug interactions. |
|---|---|
| Journal, issue, pages | Nature, Vol. 629, Issue 8010, Page 235-243, Year 2024 |
| Publish date | Mar 18, 2024 |
 Authors Authors | Jin Ye / Huaping Chen / Kaituo Wang / Yi Wang / Aaron Ammerman / Samjhana Awasthi / Jinbin Xu / Bin Liu / Weikai Li /   |
| PubMed Abstract | Biogenic monoamines-vital transmitters orchestrating neurological, endocrinal and immunological functions-are stored in secretory vesicles by vesicular monoamine transporters (VMATs) for controlled ...Biogenic monoamines-vital transmitters orchestrating neurological, endocrinal and immunological functions-are stored in secretory vesicles by vesicular monoamine transporters (VMATs) for controlled quantal release. Harnessing proton antiport, VMATs enrich monoamines around 10,000-fold and sequester neurotoxicants to protect neurons. VMATs are targeted by an arsenal of therapeutic drugs and imaging agents to treat and monitor neurodegenerative disorders, hypertension and drug addiction. However, the structural mechanisms underlying these actions remain unclear. Here we report eight cryo-electron microscopy structures of human VMAT1 in unbound form and in complex with four monoamines (dopamine, noradrenaline, serotonin and histamine), the Parkinsonism-inducing MPP, the psychostimulant amphetamine and the antihypertensive drug reserpine. Reserpine binding captures a cytoplasmic-open conformation, whereas the other structures show a lumenal-open conformation stabilized by extensive gating interactions. The favoured transition to this lumenal-open state contributes to monoamine accumulation, while protonation facilitates the cytoplasmic-open transition and concurrently prevents monoamine binding to avoid unintended depletion. Monoamines and neurotoxicants share a binding pocket that possesses polar sites for specificity and a wrist-and-fist shape for versatility. Variations in this pocket explain substrate preferences across the SLC18 family. Overall, these structural insights and supporting functional studies elucidate the mechanism of vesicular monoamine transport and provide the basis to develop therapeutics for neurodegenerative diseases and substance abuse. |
 External links External links |  Nature / Nature /  PubMed:38499039 / PubMed:38499039 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.3 - 3.7 Å |
| Structure data | EMDB-41235, PDB-8tgg: EMDB-41236, PDB-8tgh: EMDB-41237, PDB-8tgi: EMDB-41238, PDB-8tgj: EMDB-41239, PDB-8tgk: EMDB-41240, PDB-8tgl: EMDB-41241, PDB-8tgm: EMDB-41242, PDB-8tgn: |
| Chemicals | 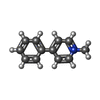 ChemComp-WRF:  ChemComp-YHR: 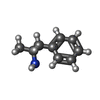 ChemComp-1WE:  ChemComp-LDP: 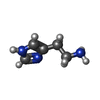 ChemComp-HSM: 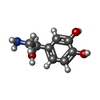 ChemComp-LNR:  ChemComp-SRO: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / VMAT / SLC18 / vascular monoamine transporter / monoamines / neurotransmitters / MPP+ / amphetamine / dopamine / reserpine / histamine / norepinephrine / serotonin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



















 homo sapiens (human)
homo sapiens (human)