+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VMAT1 dimer with histamine and reserpine | |||||||||
 Map data Map data | VMAT1 dimer with histamine and reserpine | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VMAT / SLC18 / vascular monoamine transporter / monoamines / neurotransmitters / histamine / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdopamine uptake / aminergic neurotransmitter loading into synaptic vesicle / norepinephrine uptake / monoamine:proton antiporter activity / clathrin-sculpted monoamine transport vesicle membrane / serotonin:sodium:chloride symporter activity / serotonin uptake / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters ...dopamine uptake / aminergic neurotransmitter loading into synaptic vesicle / norepinephrine uptake / monoamine:proton antiporter activity / clathrin-sculpted monoamine transport vesicle membrane / serotonin:sodium:chloride symporter activity / serotonin uptake / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters / xenobiotic transmembrane transporter activity / secretory granule membrane / terminal bouton / synaptic vesicle membrane / presynapse / endoplasmic reticulum membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Ye J / Liu B / Li W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural insights into vesicular monoamine storage and drug interactions. Authors: Jin Ye / Huaping Chen / Kaituo Wang / Yi Wang / Aaron Ammerman / Samjhana Awasthi / Jinbin Xu / Bin Liu / Weikai Li /   Abstract: Biogenic monoamines-vital transmitters orchestrating neurological, endocrinal and immunological functions-are stored in secretory vesicles by vesicular monoamine transporters (VMATs) for controlled ...Biogenic monoamines-vital transmitters orchestrating neurological, endocrinal and immunological functions-are stored in secretory vesicles by vesicular monoamine transporters (VMATs) for controlled quantal release. Harnessing proton antiport, VMATs enrich monoamines around 10,000-fold and sequester neurotoxicants to protect neurons. VMATs are targeted by an arsenal of therapeutic drugs and imaging agents to treat and monitor neurodegenerative disorders, hypertension and drug addiction. However, the structural mechanisms underlying these actions remain unclear. Here we report eight cryo-electron microscopy structures of human VMAT1 in unbound form and in complex with four monoamines (dopamine, noradrenaline, serotonin and histamine), the Parkinsonism-inducing MPP, the psychostimulant amphetamine and the antihypertensive drug reserpine. Reserpine binding captures a cytoplasmic-open conformation, whereas the other structures show a lumenal-open conformation stabilized by extensive gating interactions. The favoured transition to this lumenal-open state contributes to monoamine accumulation, while protonation facilitates the cytoplasmic-open transition and concurrently prevents monoamine binding to avoid unintended depletion. Monoamines and neurotoxicants share a binding pocket that possesses polar sites for specificity and a wrist-and-fist shape for versatility. Variations in this pocket explain substrate preferences across the SLC18 family. Overall, these structural insights and supporting functional studies elucidate the mechanism of vesicular monoamine transport and provide the basis to develop therapeutics for neurodegenerative diseases and substance abuse. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41239.map.gz emd_41239.map.gz | 109.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41239-v30.xml emd-41239-v30.xml emd-41239.xml emd-41239.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41239_fsc.xml emd_41239_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_41239.png emd_41239.png | 99.5 KB | ||
| Filedesc metadata |  emd-41239.cif.gz emd-41239.cif.gz | 6.1 KB | ||
| Others |  emd_41239_half_map_1.map.gz emd_41239_half_map_1.map.gz emd_41239_half_map_2.map.gz emd_41239_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41239 http://ftp.pdbj.org/pub/emdb/structures/EMD-41239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41239 | HTTPS FTP |
-Related structure data
| Related structure data |  8tgkMC  8tggC  8tghC  8tgiC  8tgjC  8tglC  8tgmC  8tgnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41239.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41239.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VMAT1 dimer with histamine and reserpine | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88533 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: VMAT1 dimer with histamine and reserpine
| File | emd_41239_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VMAT1 dimer with histamine and reserpine | ||||||||||||
| Projections & Slices |
| ||||||||||||
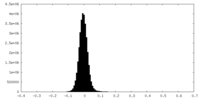
| Density Histograms |
-Half map: VMAT1 dimer with histamine and reserpine
| File | emd_41239_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VMAT1 dimer with histamine and reserpine | ||||||||||||
| Projections & Slices |
| ||||||||||||
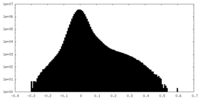
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of VMAT1 dimer with histamine and Reserpine
| Entire | Name: Complex of VMAT1 dimer with histamine and Reserpine |
|---|---|
| Components |
|
-Supramolecule #1: Complex of VMAT1 dimer with histamine and Reserpine
| Supramolecule | Name: Complex of VMAT1 dimer with histamine and Reserpine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105.2 KDa |
-Macromolecule #1: Chromaffin granule amine transporter
| Macromolecule | Name: Chromaffin granule amine transporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.565641 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MLRTILDAPQ RLLKEGRASR QLVLVVVFVA LLLDNMLFTV VVPIVPTFLY DMEFKEVNSS LHLGHAGSNC LQGTGFLEEE ITRVGVLFA SKAVMQLLVN PFVGPLTNRI GYHIPMFAGF VIMFLSTVMF AFSGTYTLLF VARTLQGIGS SFSSVAGLGM L ASVYTDDH ...String: MLRTILDAPQ RLLKEGRASR QLVLVVVFVA LLLDNMLFTV VVPIVPTFLY DMEFKEVNSS LHLGHAGSNC LQGTGFLEEE ITRVGVLFA SKAVMQLLVN PFVGPLTNRI GYHIPMFAGF VIMFLSTVMF AFSGTYTLLF VARTLQGIGS SFSSVAGLGM L ASVYTDDH ERGRAMGTAL GGLALGLLVG APFGSVMYEF VGKSAPFLIL AFLALLDGAL QLCILQPSKV SPESAKGTPL FM LLKDPYI LVAAGSICFA NMGVAILEPT LPIWMMQTMC SPKWQLGLAF LPASVSYLIG TNLFGVLANK MGRWLCSLIG MLV VGTSLL CVPLAHNIFG LIGPNAGLGL AIGMVDSSMM PIMGHLVDLR HTSVYGSVYA IADVAFCMGF AIGPSTGGAI VKAI GFPWL MVITGVINIV YAPLCYYLRS PPAKEEKLAI LSQDCPMETR MYATQKPTKE FPLGEDSDEE PDHEE UniProtKB: Chromaffin granule amine transporter, Chromaffin granule amine transporter |
-Macromolecule #2: HISTAMINE
| Macromolecule | Name: HISTAMINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: HSM |
|---|---|
| Molecular weight | Theoretical: 111.145 Da |
| Chemical component information |  ChemComp-HSM: |
-Macromolecule #3: reserpine
| Macromolecule | Name: reserpine / type: ligand / ID: 3 / Number of copies: 1 / Formula: YHR |
|---|---|
| Molecular weight | Theoretical: 608.679 Da |
| Chemical component information |  ChemComp-YHR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 63.0 K / Max: 77.2 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6166 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)