+Search query
-Structure paper
| Title | Structural basis for translation inhibition by MERS-CoV Nsp1 reveals a conserved mechanism for betacoronaviruses. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 42, Issue 10, Page 113156, Year 2023 |
| Publish date | Sep 19, 2023 |
 Authors Authors | Swapnil C Devarkar / Michael Vetick / Shravani Balaji / Ivan B Lomakin / Luojia Yang / Danni Jin / Wendy V Gilbert / Sidi Chen / Yong Xiong /  |
| PubMed Abstract | All betacoronaviruses (β-CoVs) encode non-structural protein 1 (Nsp1), an essential pathogenicity factor that potently restricts host gene expression. Among the β-CoV family, MERS-CoV is the most ...All betacoronaviruses (β-CoVs) encode non-structural protein 1 (Nsp1), an essential pathogenicity factor that potently restricts host gene expression. Among the β-CoV family, MERS-CoV is the most distantly related member to SARS-CoV-2, and the mechanism for host translation inhibition by MERS-CoV Nsp1 remains controversial. Herein, we show that MERS-CoV Nsp1 directly interacts with the 40S ribosomal subunit. Using cryogenic electron microscopy (cryo-EM), we report a 2.6-Å structure of the MERS-CoV Nsp1 bound to the human 40S ribosomal subunit. The extensive interactions between C-terminal domain of MERS-CoV Nsp1 and the mRNA entry channel of the 40S ribosomal subunit are critical for its translation inhibition function. This mechanism of MERS-CoV Nsp1 is strikingly similar to SARS-CoV and SARS-CoV-2 Nsp1, despite modest sequence conservation. Our results reveal that the mechanism of host translation inhibition is conserved across β-CoVs and highlight a potential therapeutic target for the development of antivirals that broadly restrict β-CoVs. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:37733586 PubMed:37733586 |
| Methods | EM (single particle) |
| Resolution | 2.6 Å |
| Structure data | EMDB-41039, PDB-8t4s:  EMDB-41063: MERS-CoV Nsp1 protein binds the human 40S ribosome - Consensus Reconstruction 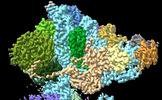 EMDB-41064: MERS-CoV Nsp1 protein bound to the human 40S ribosome: Focus Refined 40S Head Map 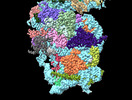 EMDB-41065: MERS-CoV Nsp1 protein bound to human 40S ribosome: 40S Body Focus Refined Map |
| Chemicals |  ChemComp-MG:  ChemComp-ZN:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | Ribosome/Viral Protein / MERS-CoV Nsp1 / translation inhibition / 40S ribosome / betacoronaviruses / RIBOSOME / Ribosome-Viral Protein complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



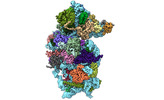

 homo sapiens (human)
homo sapiens (human)
