+Search query
-Structure paper
| Title | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. |
|---|---|
| Journal, issue, pages | Nat Chem Biol, Vol. 16, Issue 1, Page 7-14, Year 2020 |
| Publish date | Nov 4, 2019 |
 Authors Authors | Tyler B Faust / Hojong Yoon / Radosław P Nowak / Katherine A Donovan / Zhengnian Li / Quan Cai / Nicholas A Eleuteri / Tinghu Zhang / Nathanael S Gray / Eric S Fischer /  |
| PubMed Abstract | The investigational drugs E7820, indisulam and tasisulam (aryl-sulfonamides) promote the degradation of the splicing factor RBM39 in a proteasome-dependent mechanism. While the activity critically ...The investigational drugs E7820, indisulam and tasisulam (aryl-sulfonamides) promote the degradation of the splicing factor RBM39 in a proteasome-dependent mechanism. While the activity critically depends on the cullin RING ligase substrate receptor DCAF15, the molecular details remain elusive. Here we present the cryo-EM structure of the DDB1-DCAF15-DDA1 core ligase complex bound to RBM39 and E7820 at a resolution of 4.4 Å, together with crystal structures of engineered subcomplexes. We show that DCAF15 adopts a new fold stabilized by DDA1, and that extensive protein-protein contacts between the ligase and substrate mitigate low affinity interactions between aryl-sulfonamides and DCAF15. Our data demonstrate how aryl-sulfonamides neo-functionalize a shallow, non-conserved pocket on DCAF15 to selectively bind and degrade RBM39 and the closely related splicing factor RBM23 without the requirement for a high-affinity ligand, which has broad implications for the de novo discovery of molecular glue degraders. |
 External links External links |  Nat Chem Biol / Nat Chem Biol /  PubMed:31686031 / PubMed:31686031 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.9 - 10.0 Å |
| Structure data |  EMDB-20553:  EMDB-20554:  PDB-6q0r:  PDB-6q0v:  PDB-6q0w: |
| Chemicals |  ChemComp-OXM: 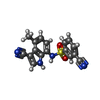 ChemComp-O6M:  ChemComp-ZN:  ChemComp-HOH: 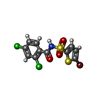 ChemComp-P7M: 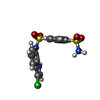 ChemComp-EF6: |
| Source |
|
 Keywords Keywords | LIGASE / Ubiquitin / homeostasis / targeted protein degradation |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 homo sapiens (human)
homo sapiens (human)