+Search query
-Structure paper
| Title | T cell toxicity induced by tigecycline binding to the mitochondrial ribosome. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 16, Issue 1, Page 4080, Year 2025 |
| Publish date | May 1, 2025 |
 Authors Authors | Qiuya Shao / Anas Khawaja / Minh Duc Nguyen / Vivek Singh / Jingdian Zhang / Yong Liu / Joel Nordin / Monika Adori / C Axel Innis / Xaquin Castro Dopico / Joanna Rorbach /      |
| PubMed Abstract | Tetracyclines are essential bacterial protein synthesis inhibitors under continual development to combat antibiotic resistance yet suffer from unwanted side effects. Mitoribosomes - responsible for ...Tetracyclines are essential bacterial protein synthesis inhibitors under continual development to combat antibiotic resistance yet suffer from unwanted side effects. Mitoribosomes - responsible for generating oxidative phosphorylation (OXPHOS) subunits - share structural similarities with bacterial machinery and may suffer from cross-reactivity. Since lymphocytes rely upon OXPHOS upregulation to establish immunity, we set out to assess the impact of ribosome-targeting antibiotics on human T cells. We find tigecycline, a third-generation tetracycline, to be the most cytotoxic compound tested. In vitro, 5-10 μM tigecycline inhibits mitochondrial but not cytosolic translation, mitochondrial complex I, III and IV expression, and curtails the activation and expansion of unique T cell subsets. By cryo-EM, we find tigecycline to occupy three sites on T cell mitoribosomes. In addition to the conserved A-site found in bacteria, tigecycline also attaches to the peptidyl transferase center of the large subunit. Furthermore, a third, distinct binding site on the large subunit, aligns with helices analogous to those in bacteria, albeit lacking methylation in humans. The data provide a mechanism to explain part of the anti-inflammatory effects of these drugs and inform antibiotic design. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:40312422 / PubMed:40312422 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.2 - 2.9 Å |
| Structure data | EMDB-19460, PDB-8rri:  EMDB-19490: Human mitoribosome Class P-tRNA, mtSSU head (local filter) with tigecycline  EMDB-19491: Human mitoribosome Class P-tRNA, mtLSU body (local filter) with tigecycline bound to mtLSU site-1 and 2  EMDB-19493: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class P-tRNA consensus.  EMDB-19526: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class A-tRNA P-tRNA consensus.  EMDB-19539: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class A-tRNA P-tRNA mtSSU head (local-filter).  EMDB-19542: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class A-tRNA P-tRNA mtLSU body (local-filter).  EMDB-19544: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class empty consensus (local-filter)  EMDB-19545: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class empty mtSSU head (local-filter)  EMDB-19546: Human mitochondrial ribosome in complex with antibiotic tigecycline, Class empty mtLSU body (local-filter) |
| Chemicals |  ChemComp-VAL:  ChemComp-MG:  ChemComp-FES:  ChemComp-ATP:  ChemComp-GDP: 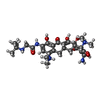 ChemComp-T1C:  ChemComp-K:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | RIBOSOME / antibiotics; immunometabolism; mitochondrial ribosomes; tetracyclines; T cells. |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)