+Search query
-Structure paper
| Title | Deciphering the allosteric regulation of mycobacterial inosine-5'-monophosphate dehydrogenase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 6673, Year 2024 |
| Publish date | Aug 6, 2024 |
 Authors Authors | Ondřej Bulvas / Zdeněk Knejzlík / Jakub Sýs / Anatolij Filimoněnko / Monika Čížková / Kamila Clarová / Dominik Rejman / Tomáš Kouba / Iva Pichová /  |
| PubMed Abstract | Allosteric regulation of inosine 5'-monophosphate dehydrogenase (IMPDH), an essential enzyme of purine metabolism, contributes to the homeostasis of adenine and guanine nucleotides. However, the ...Allosteric regulation of inosine 5'-monophosphate dehydrogenase (IMPDH), an essential enzyme of purine metabolism, contributes to the homeostasis of adenine and guanine nucleotides. However, the precise molecular mechanism of IMPDH regulation in bacteria remains unclear. Using biochemical and cryo-EM approaches, we reveal the intricate molecular mechanism of the IMPDH allosteric regulation in mycobacteria. The enzyme is inhibited by both GTP and (p)ppGpp, which bind to the regulatory CBS domains and, via interactions with basic residues in hinge regions, lock the catalytic core domains in a compressed conformation. This results in occlusion of inosine monophosphate (IMP) substrate binding to the active site and, ultimately, inhibition of the enzyme. The GTP and (p)ppGpp allosteric effectors bind to their dedicated sites but stabilize the compressed octamer by a common mechanism. Inhibition is relieved by the competitive displacement of GTP or (p)ppGpp by ATP allowing IMP-induced enzyme expansion. The structural knowledge and mechanistic understanding presented here open up new possibilities for the development of allosteric inhibitors with antibacterial potential. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:39107302 / PubMed:39107302 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.35 - 3.27 Å |
| Structure data | EMDB-17988, PDB-8pw3: EMDB-18184, PDB-8q65: EMDB-18600: Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+GTP-bound form, compressed EMDB-18601: Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+GTP-bound form, less-compressed EMDB-18602: Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+ppGpp-bound form, compressed EMDB-18604: Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+ppGpp-bound form, less-compressed EMDB-18606, PDB-8qqv: EMDB-18607, PDB-8qqw: EMDB-18608, PDB-8qqx: |
| Chemicals |  ChemComp-ATP:  ChemComp-MG:  ChemComp-GTP:  ChemComp-HOH: 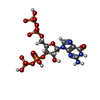 ChemComp-G4P: 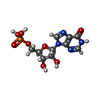 ChemComp-IMP: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / Octamer / Apo enzyme / Purine metabolism / IMPDH / ATP-bound complex / ATP+GTP complex / ATP+ppGpp complex / ATP+IMP complex / ATP complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





















 mycolicibacterium smegmatis mc2 155 (bacteria)
mycolicibacterium smegmatis mc2 155 (bacteria)