+Search query
-Structure paper
| Title | The respiratory supercomplex from C. glutamicum. |
|---|---|
| Journal, issue, pages | Structure, Vol. 30, Issue 3, Page 338-349.e3, Year 2022 |
| Publish date | Mar 3, 2022 |
 Authors Authors | Agnes Moe / Terezia Kovalova / Sylwia Król / David J Yanofsky / Michael Bott / Dan Sjöstrand / John L Rubinstein / Martin Högbom / Peter Brzezinski /    |
| PubMed Abstract | Corynebacterium glutamicum is a preferentially aerobic gram-positive bacterium belonging to the phylum Actinobacteria, which also includes the pathogen Mycobacterium tuberculosis. In these bacteria, ...Corynebacterium glutamicum is a preferentially aerobic gram-positive bacterium belonging to the phylum Actinobacteria, which also includes the pathogen Mycobacterium tuberculosis. In these bacteria, respiratory complexes III and IV form a CIIICIV supercomplex that catalyzes oxidation of menaquinol and reduction of dioxygen to water. We isolated the C. glutamicum supercomplex and used cryo-EM to determine its structure at 2.9 Å resolution. The structure shows a central CIII dimer flanked by a CIV on two sides. A menaquinone is bound in each of the Q and Q sites in each CIII and an additional menaquinone is positioned ∼14 Å from heme b. A di-heme cyt. cc subunit electronically connects each CIII with an adjacent CIV, with the Rieske iron-sulfur protein positioned with the iron near heme b. Multiple subunits interact to form a convoluted sub-structure at the cytoplasmic side of the supercomplex, which defines a path for proton transfer into CIV. |
 External links External links |  Structure / Structure /  PubMed:34910901 PubMed:34910901 |
| Methods | EM (single particle) |
| Resolution | 2.9 Å |
| Structure data | EMDB-13777, PDB-7q21: |
| Chemicals |  ChemComp-CDL:  ChemComp-7PH:  ChemComp-TRD:  ChemComp-9XX:  ChemComp-TWT:  ChemComp-CU:  ChemComp-MG: 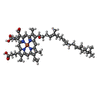 ChemComp-HAS:  ChemComp-CA:  ChemComp-FES:  ChemComp-MQ9:  ChemComp-9YF:  ChemComp-PLM:  ChemComp-HEM:  ChemComp-HEC: |
| Source |
|
 Keywords Keywords | ELECTRON TRANSPORT / MEMBRANE PROTEIN / CRYO-EM / RESPIRATORY SUPERCOMPLEX / ACTINOBACTERIA |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 corynebacterium glutamicum atcc 13032 (bacteria)
corynebacterium glutamicum atcc 13032 (bacteria)