[English] 日本語
 Yorodumi
Yorodumi- EMDB-13777: III2-IV2 respiratory supercomplex from Corynebacterium glutamicum -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13777 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / CRYO-EM / RESPIRATORY SUPERCOMPLEX / ACTINOBACTERIA / ELECTRON TRANSPORT | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain ...aerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain / monooxygenase activity / electron transport chain / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Corynebacterium glutamicum ATCC 13032 (bacteria) / Corynebacterium glutamicum ATCC 13032 (bacteria) /  Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) (bacteria) Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||
 Authors Authors | Kovalova T / Moe A | |||||||||||||||
| Funding support |  Canada, 4 items Canada, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: The respiratory supercomplex from C. glutamicum. Authors: Agnes Moe / Terezia Kovalova / Sylwia Król / David J Yanofsky / Michael Bott / Dan Sjöstrand / John L Rubinstein / Martin Högbom / Peter Brzezinski /    Abstract: Corynebacterium glutamicum is a preferentially aerobic gram-positive bacterium belonging to the phylum Actinobacteria, which also includes the pathogen Mycobacterium tuberculosis. In these bacteria, ...Corynebacterium glutamicum is a preferentially aerobic gram-positive bacterium belonging to the phylum Actinobacteria, which also includes the pathogen Mycobacterium tuberculosis. In these bacteria, respiratory complexes III and IV form a CIIICIV supercomplex that catalyzes oxidation of menaquinol and reduction of dioxygen to water. We isolated the C. glutamicum supercomplex and used cryo-EM to determine its structure at 2.9 Å resolution. The structure shows a central CIII dimer flanked by a CIV on two sides. A menaquinone is bound in each of the Q and Q sites in each CIII and an additional menaquinone is positioned ∼14 Å from heme b. A di-heme cyt. cc subunit electronically connects each CIII with an adjacent CIV, with the Rieske iron-sulfur protein positioned with the iron near heme b. Multiple subunits interact to form a convoluted sub-structure at the cytoplasmic side of the supercomplex, which defines a path for proton transfer into CIV. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13777.map.gz emd_13777.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13777-v30.xml emd-13777-v30.xml emd-13777.xml emd-13777.xml | 35.3 KB 35.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13777.png emd_13777.png | 89.2 KB | ||
| Filedesc metadata |  emd-13777.cif.gz emd-13777.cif.gz | 9.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13777 http://ftp.pdbj.org/pub/emdb/structures/EMD-13777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13777 | HTTPS FTP |
-Related structure data
| Related structure data |  7q21MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13777.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13777.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
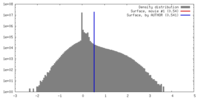
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : The respiratory supercomplex
+Supramolecule #1: The respiratory supercomplex
+Macromolecule #1: Co-purified unknown transmembrane helices built as polyALA (AscD)
+Macromolecule #2: Cytochrome c oxidase subunit 3
+Macromolecule #3: Uncharacterized membrane protein Cgl2017/cg2211
+Macromolecule #4: Cytochrome c oxidase polypeptide 4
+Macromolecule #5: Cytochrome c oxidase subunit 2
+Macromolecule #6: Cytochrome c oxidase subunit 1
+Macromolecule #7: Cytochrome bc1 complex Rieske iron-sulfur subunit
+Macromolecule #8: Uncharacterized protein Cgl2664/cg2949
+Macromolecule #9: Cytochrome bc1 complex cytochrome b subunit
+Macromolecule #10: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #11: Co-purified unknown peptide built as polyALA (AscE)
+Macromolecule #12: Co-purified unknown peptide built as polyALA (AscE)
+Macromolecule #13: Actinobacterial supercomplex, subunit C (AscC)
+Macromolecule #14: Hypothetical membrane protein
+Macromolecule #15: CARDIOLIPIN
+Macromolecule #16: (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate
+Macromolecule #17: TRIDECANE
+Macromolecule #18: (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate
+Macromolecule #19: DOCOSANE
+Macromolecule #20: COPPER (II) ION
+Macromolecule #21: MAGNESIUM ION
+Macromolecule #22: HEME-AS
+Macromolecule #23: CALCIUM ION
+Macromolecule #24: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #25: MENAQUINONE-9
+Macromolecule #26: (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3...
+Macromolecule #27: PALMITIC ACID
+Macromolecule #28: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #29: HEME C
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 276 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 8.0 sec. / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 65391 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)