+Search query
-Structure paper
| Title | Structural basis of phosphatidylinositol 3-kinase C2α function. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 29, Issue 3, Page 218-228, Year 2022 |
| Publish date | Mar 7, 2022 |
 Authors Authors | Wen-Ting Lo / Yingyi Zhang / Oscar Vadas / Yvette Roske / Federico Gulluni / Maria Chiara De Santis / Andreja Vujicic Zagar / Heike Stephanowitz / Emilio Hirsch / Fan Liu / Oliver Daumke / Misha Kudryashev / Volker Haucke /     |
| PubMed Abstract | Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, ...Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, viral replication, platelet formation and a role in mitosis. The molecular basis of these activities of PI3KC2α is poorly understood. Here, we report high-resolution crystal structures as well as a 4.4-Å cryogenic-electron microscopic (cryo-EM) structure of PI3KC2α in active and inactive conformations. We unravel a coincident mechanism of lipid-induced activation of PI3KC2α at membranes that involves large-scale repositioning of its Ras-binding and lipid-binding distal Phox-homology and C-C2 domains, and can serve as a model for the entire class II PI3K family. Moreover, we describe a PI3KC2α-specific helical bundle domain that underlies its scaffolding function at the mitotic spindle. Our results advance our understanding of PI3K biology and pave the way for the development of specific inhibitors of class II PI3K function with wide applications in biomedicine. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:35256802 / PubMed:35256802 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.42 - 4.4 Å |
| Structure data |  EMDB-12191:  PDB-7bi2:  PDB-7bi4:  PDB-7bi6:  PDB-7bi9: |
| Chemicals |  ChemComp-EDO: 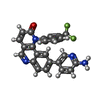 ChemComp-17G:  ChemComp-IOD:  ChemComp-HOH:  ChemComp-SO4:  ChemComp-ATP:  ChemComp-MG: 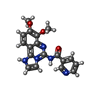 ChemComp-090: |
| Source |
|
 Keywords Keywords | TRANSFERASE / PI3KC2 alpha / kinase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




