+Search query
-Structure paper
| Title | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis. |
|---|---|
| Journal, issue, pages | Cell, Vol. 187, Issue 13, Page 3303-3318.e18, Year 2024 |
| Publish date | Jun 20, 2024 |
 Authors Authors | Jannik Hugener / Jingwei Xu / Rahel Wettstein / Lydia Ioannidi / Daniel Velikov / Florian Wollweber / Adrian Henggeler / Joao Matos / Martin Pilhofer /   |
| PubMed Abstract | Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly ...Gamete formation and subsequent offspring development often involve extended phases of suspended cellular development or even dormancy. How cells adapt to recover and resume growth remains poorly understood. Here, we visualized budding yeast cells undergoing meiosis by cryo-electron tomography (cryoET) and discovered elaborate filamentous assemblies decorating the nucleus, cytoplasm, and mitochondria. To determine filament composition, we developed a "filament identification" (FilamentID) workflow that combines multiscale cryoET/cryo-electron microscopy (cryoEM) analyses of partially lysed cells or organelles. FilamentID identified the mitochondrial filaments as being composed of the conserved aldehyde dehydrogenase Ald4 and the nucleoplasmic/cytoplasmic filaments as consisting of acetyl-coenzyme A (CoA) synthetase Acs1. Structural characterization further revealed the mechanism underlying polymerization and enabled us to genetically perturb filament formation. Acs1 polymerization facilitates the recovery of chronologically aged spores and, more generally, the cell cycle re-entry of starved cells. FilamentID is broadly applicable to characterize filaments of unknown identity in diverse cellular contexts. |
 External links External links |  Cell / Cell /  PubMed:38906101 PubMed:38906101 |
| Methods | EM (helical sym.) / EM (single particle) / EM (subtomogram averaging) / EM (tomography) |
| Resolution | 3.5 - 44.0 Å |
| Structure data | EMDB-19548, PDB-8rwj: EMDB-19549, PDB-8rwk:  EMDB-19550: cryoEM structure of purified Acs1 filament from meiotic yeast cells 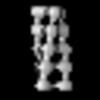 EMDB-19551: cryo sub-tomogram average of Acs1 filament from spread meiotic yeast spheroplasts 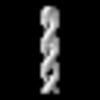 EMDB-19552: cryo sub-tomogram average of Ald4 filaments from purified meiotic yeast mitochondria  EMDB-19553: cryo-tomogram of FIB-milled meiotic yeast cell containing filaments in mitochondria  EMDB-19554: cryo-tomogram of FIB-milled meiotic yeast cell containing filaments in the nucleus 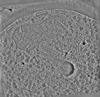 EMDB-19555: cryo-tomogram of FIB-milled meiotic yeast cell containing filaments in the cytoplasm  EMDB-19556: cryo-tomogram of purified meiotic yeast mitochondrion containing Ald4 filaments  EMDB-19557: cryo-tomogram of spread meiotic yeast spheroplast containing Acs1 filaments  EMDB-19558: cryo-tomogram of spread starved yeast spheroplast containing filaments |
| Chemicals | 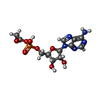 ChemComp-6R9:  ChemComp-NAP: |
| Source |
|
 Keywords Keywords | CYTOSOLIC PROTEIN / metabolic enzyme / filament / cryoEM |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








