+Search query
-Structure paper
| Title | Ligand recognition and G-protein coupling of trace amine receptor TAAR1. |
|---|---|
| Journal, issue, pages | Nature, Vol. 624, Issue 7992, Page 672-681, Year 2023 |
| Publish date | Nov 7, 2023 |
 Authors Authors | Zheng Xu / Lulu Guo / Jingjing Yu / Siyuan Shen / Chao Wu / Weifeng Zhang / Chang Zhao / Yue Deng / Xiaowen Tian / Yuying Feng / Hanlin Hou / Lantian Su / Hongshuang Wang / Shuo Guo / Heli Wang / Kexin Wang / Peipei Chen / Jie Zhao / Xiaoyu Zhang / Xihao Yong / Lin Cheng / Lunxu Liu / Shengyong Yang / Fan Yang / Xiaohui Wang / Xiao Yu / Yunfei Xu / Jin-Peng Sun / Wei Yan / Zhenhua Shao /  |
| PubMed Abstract | Trace-amine-associated receptors (TAARs), a group of biogenic amine receptors, have essential roles in neurological and metabolic homeostasis. They recognize diverse endogenous trace amines and ...Trace-amine-associated receptors (TAARs), a group of biogenic amine receptors, have essential roles in neurological and metabolic homeostasis. They recognize diverse endogenous trace amines and subsequently activate a range of G-protein-subtype signalling pathways. Notably, TAAR1 has emerged as a promising therapeutic target for treating psychiatric disorders. However, the molecular mechanisms underlying its ability to recognize different ligands remain largely unclear. Here we present nine cryo-electron microscopy structures, with eight showing human and mouse TAAR1 in a complex with an array of ligands, including the endogenous 3-iodothyronamine, two antipsychotic agents, the psychoactive drug amphetamine and two identified catecholamine agonists, and one showing 5-HTR in a complex with an antipsychotic agent. These structures reveal a rigid consensus binding motif in TAAR1 that binds to endogenous trace amine stimuli and two extended binding pockets that accommodate diverse chemotypes. Combined with mutational analysis, functional assays and molecular dynamic simulations, we elucidate the structural basis of drug polypharmacology and identify the species-specific differences between human and mouse TAAR1. Our study provides insights into the mechanism of ligand recognition and G-protein selectivity by TAAR1, which may help in the discovery of ligands or therapeutic strategies for neurological and metabolic disorders. |
 External links External links |  Nature / Nature /  PubMed:37935376 PubMed:37935376 |
| Methods | EM (single particle) |
| Resolution | 2.84 - 3.65 Å |
| Structure data | EMDB-36399, PDB-8jlj: EMDB-36400, PDB-8jlk: EMDB-36401, PDB-8jln: EMDB-36402, PDB-8jlo: EMDB-36403, PDB-8jlp: EMDB-36404, PDB-8jlq: EMDB-36405, PDB-8jlr: EMDB-36625, PDB-8jso: EMDB-36626, PDB-8jsp: |
| Chemicals | 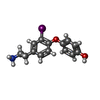 ChemComp-UJF:  ChemComp-UJL: 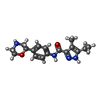 ChemComp-UJU:  ChemComp-G3C: 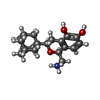 ChemComp-VRK: 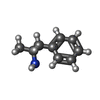 ChemComp-1WE: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN/IMMUNE SYSTEM / Complex / Agonist / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





















 homo sapiens (human)
homo sapiens (human)

