+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fenoldopam-bound hTAAR1-Gs protein complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Agonist / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationAmine ligand-binding receptors / trace-amine receptor activity / endomembrane system / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta ...Amine ligand-binding receptors / trace-amine receptor activity / endomembrane system / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / endoplasmic reticulum membrane / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Xu Z / Guo LL / Zhao C / Shen SY / Sun JP / Shao ZH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Ligand recognition and G-protein coupling of trace amine receptor TAAR1. Authors: Zheng Xu / Lulu Guo / Jingjing Yu / Siyuan Shen / Chao Wu / Weifeng Zhang / Chang Zhao / Yue Deng / Xiaowen Tian / Yuying Feng / Hanlin Hou / Lantian Su / Hongshuang Wang / Shuo Guo / Heli ...Authors: Zheng Xu / Lulu Guo / Jingjing Yu / Siyuan Shen / Chao Wu / Weifeng Zhang / Chang Zhao / Yue Deng / Xiaowen Tian / Yuying Feng / Hanlin Hou / Lantian Su / Hongshuang Wang / Shuo Guo / Heli Wang / Kexin Wang / Peipei Chen / Jie Zhao / Xiaoyu Zhang / Xihao Yong / Lin Cheng / Lunxu Liu / Shengyong Yang / Fan Yang / Xiaohui Wang / Xiao Yu / Yunfei Xu / Jin-Peng Sun / Wei Yan / Zhenhua Shao /  Abstract: Trace-amine-associated receptors (TAARs), a group of biogenic amine receptors, have essential roles in neurological and metabolic homeostasis. They recognize diverse endogenous trace amines and ...Trace-amine-associated receptors (TAARs), a group of biogenic amine receptors, have essential roles in neurological and metabolic homeostasis. They recognize diverse endogenous trace amines and subsequently activate a range of G-protein-subtype signalling pathways. Notably, TAAR1 has emerged as a promising therapeutic target for treating psychiatric disorders. However, the molecular mechanisms underlying its ability to recognize different ligands remain largely unclear. Here we present nine cryo-electron microscopy structures, with eight showing human and mouse TAAR1 in a complex with an array of ligands, including the endogenous 3-iodothyronamine, two antipsychotic agents, the psychoactive drug amphetamine and two identified catecholamine agonists, and one showing 5-HTR in a complex with an antipsychotic agent. These structures reveal a rigid consensus binding motif in TAAR1 that binds to endogenous trace amine stimuli and two extended binding pockets that accommodate diverse chemotypes. Combined with mutational analysis, functional assays and molecular dynamic simulations, we elucidate the structural basis of drug polypharmacology and identify the species-specific differences between human and mouse TAAR1. Our study provides insights into the mechanism of ligand recognition and G-protein selectivity by TAAR1, which may help in the discovery of ligands or therapeutic strategies for neurological and metabolic disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36404.map.gz emd_36404.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36404-v30.xml emd-36404-v30.xml emd-36404.xml emd-36404.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36404.png emd_36404.png | 29.5 KB | ||
| Filedesc metadata |  emd-36404.cif.gz emd-36404.cif.gz | 7.5 KB | ||
| Others |  emd_36404_half_map_1.map.gz emd_36404_half_map_1.map.gz emd_36404_half_map_2.map.gz emd_36404_half_map_2.map.gz | 31.7 MB 31.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36404 http://ftp.pdbj.org/pub/emdb/structures/EMD-36404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36404 | HTTPS FTP |
-Related structure data
| Related structure data |  8jlqMC  8jljC  8jlkC  8jlnC  8jloC  8jlpC  8jlrC  8jsoC  8jspC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36404.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36404.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
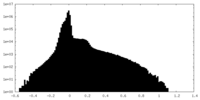
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
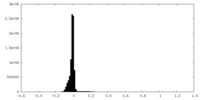
-Half map: #2
| File | emd_36404_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
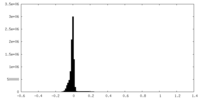
-Half map: #1
| File | emd_36404_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : hTAAR1-bound T1AM in complex with Gs heterotrimer
+Supramolecule #1: hTAAR1-bound T1AM in complex with Gs heterotrimer
+Supramolecule #2: Gs heterotrimer
+Supramolecule #3: scFV16
+Supramolecule #4: T1AM-bound hTAAR1
+Macromolecule #1: mGs
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Soluble cytochrome b562,Trace amine-associated receptor 1
+Macromolecule #4: ScFv16
+Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #6: (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benz...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)