+Search query
-Structure paper
| Title | Structural basis of amine odorant perception by a mammal olfactory receptor. |
|---|---|
| Journal, issue, pages | Nature, Vol. 618, Issue 7963, Page 193-200, Year 2023 |
| Publish date | May 24, 2023 |
 Authors Authors | Lulu Guo / Jie Cheng / Shuo Lian / Qun Liu / Yan Lu / Yuan Zheng / Kongkai Zhu / Minghui Zhang / Yalei Kong / Chao Zhang / Naikang Rong / Yuming Zhuang / Guoxing Fang / Jingjing Jiang / Tianyao Zhang / Xiang Han / Zili Liu / Ming Xia / Shangming Liu / Lei Zhang / Stephen D Liberles / Xiao Yu / Yunfei Xu / Fan Yang / Qian Li / Jin-Peng Sun /   |
| PubMed Abstract | Odorants are detected as smell in the nasal epithelium of mammals by two G-protein-coupled receptor families, the odorant receptors and the trace amine-associated receptors (TAARs). TAARs emerged ...Odorants are detected as smell in the nasal epithelium of mammals by two G-protein-coupled receptor families, the odorant receptors and the trace amine-associated receptors (TAARs). TAARs emerged following the divergence of jawed and jawless fish, and comprise a large monophyletic family of receptors that recognize volatile amine odorants to elicit both intraspecific and interspecific innate behaviours such as attraction and aversion. Here we report cryo-electron microscopy structures of mouse TAAR9 (mTAAR9) and mTAAR9-G or mTAAR9-G trimers in complex with β-phenylethylamine, N,N-dimethylcyclohexylamine or spermidine. The mTAAR9 structures contain a deep and tight ligand-binding pocket decorated with a conserved DWY motif, which is essential for amine odorant recognition. In the mTAAR9 structure, a unique disulfide bond connecting the N terminus to ECL2 is required for agonist-induced receptor activation. We identify key structural motifs of TAAR family members for detecting monoamines and polyamines and the shared sequence of different TAAR members that are responsible for recognition of the same odour chemical. We elucidate the molecular basis of mTAAR9 coupling to G and G by structural characterization and mutational analysis. Collectively, our results provide a structural basis for odorant detection, receptor activation and G coupling of an amine olfactory receptor. |
 External links External links |  Nature / Nature /  PubMed:37225986 PubMed:37225986 |
| Methods | EM (single particle) |
| Resolution | 2.97 - 3.49 Å |
| Structure data | EMDB-35705, PDB-8itf: EMDB-35761, PDB-8iw1: EMDB-35762, PDB-8iw4: EMDB-35763, PDB-8iw7: EMDB-35764, PDB-8iw9: EMDB-35765, PDB-8iwe: EMDB-35771, PDB-8iwm: |
| Chemicals | 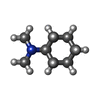 ChemComp-8IA:  ChemComp-SPD: 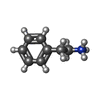 ChemComp-PEA: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / DMCHA / mTAAR9 / PEA / SPE / CAD |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers


















 homo sapiens (human)
homo sapiens (human)
