+Search query
-Structure paper
| Title | Structure and molecular assignment of lactococcal phage TP901-1 baseplate. |
|---|---|
| Journal, issue, pages | J Biol Chem, Vol. 285, Issue 50, Page 39079-39086, Year 2010 |
| Publish date | Dec 10, 2010 |
 Authors Authors | Cecilia Bebeacua / Patrick Bron / Livia Lai / Christina Skovgaard Vegge / Lone Brøndsted / Silvia Spinelli / Valérie Campanacci / David Veesler / Marin van Heel / Christian Cambillau /  |
| PubMed Abstract | P335 lactococcal phages infect the gram(+) bacterium Lactococcus lactis using a large multiprotein complex located at the distal part of the tail and termed baseplate (BP). The BP harbors the ...P335 lactococcal phages infect the gram(+) bacterium Lactococcus lactis using a large multiprotein complex located at the distal part of the tail and termed baseplate (BP). The BP harbors the receptor-binding proteins (RBPs), which allow the specific recognition of saccharidic receptors localized on the host cell surface. We report here the electron microscopic structure of the phage TP901-1 wild-type BP as well as those of two mutants bppL (-) and bppU(-), lacking BppL (the RBPs) or both peripheral BP components (BppL and BppU), respectively. We also achieved an electron microscopic reconstruction of a partial BP complex, formed by BppU and BppL. This complex exhibits a tripod shape and is composed of nine BppLs and three BppUs. These structures, combined with light-scattering measurements, led us to propose that the TP901-1 BP harbors six tripods at its periphery, located around the central tube formed by ORF46 (Dit) hexamers, at its proximal end, and a ORF47 (Tal) trimer at its distal extremity. A total of 54 BppLs (18 RBPs) are thus available to mediate host anchoring with a large apparent avidity. TP901-1 BP exhibits an infection-ready conformation and differs strikingly from the lactococcal phage p2 BP, bearing only 6 RBPs, and which needs a conformational change to reach its activated state. The comparison of several Siphoviridae structures uncovers a close organization of their central BP core whereas striking differences occur at the periphery, leading to diverse mechanisms of host recognition. |
 External links External links |  J Biol Chem / J Biol Chem /  PubMed:20937834 / PubMed:20937834 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.85 - 28.0 Å |
| Structure data | 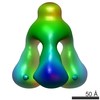 EMDB-1792:  EMDB-1793: 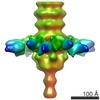 EMDB-1794:  EMDB-1795:  PDB-3ejc: |
| Chemicals |  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | VIRAL PROTEIN / SUGAR BINDING PROTEIN / Lactococcus lactis / siphoviridae / receptor binding protein / phage TP901-1 / P335 species |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 lactococcus phage tp901-1 (virus)
lactococcus phage tp901-1 (virus)