+Search query
-Structure paper
| Title | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate. |
|---|---|
| Journal, issue, pages | Cell, Vol. 186, Issue 12, Page 2644-2655.e16, Year 2023 |
| Publish date | Jun 8, 2023 |
 Authors Authors | Hongwen Chen / Shahbaz Ahmed / Hongtu Zhao / Nadia Elghobashi-Meinhardt / Yaxin Dai / Jae Hun Kim / Jeffrey G McDonald / Xiaochun Li / Chia-Hsueh Lee /   |
| PubMed Abstract | Sphingosine-1-phosphate (S1P) is an important signaling sphingolipid that regulates the immune system, angiogenesis, auditory function, and epithelial and endothelial barrier integrity. Spinster ...Sphingosine-1-phosphate (S1P) is an important signaling sphingolipid that regulates the immune system, angiogenesis, auditory function, and epithelial and endothelial barrier integrity. Spinster homolog 2 (Spns2) is an S1P transporter that exports S1P to initiate lipid signaling cascades. Modulating Spns2 activity can be beneficial in treatments of cancer, inflammation, and immune diseases. However, the transport mechanism of Spns2 and its inhibition remain unclear. Here, we present six cryo-EM structures of human Spns2 in lipid nanodiscs, including two functionally relevant intermediate conformations that link the inward- and outward-facing states, to reveal the structural basis of the S1P transport cycle. Functional analyses suggest that Spns2 exports S1P via facilitated diffusion, a mechanism distinct from other MFS lipid transporters. Finally, we show that the Spns2 inhibitor 16d attenuates the transport activity by locking Spns2 in the inward-facing state. Our work sheds light on Spns2-mediated S1P transport and aids the development of advanced Spns2 inhibitors. |
 External links External links |  Cell / Cell /  PubMed:37224812 / PubMed:37224812 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.93 - 4.17 Å |
| Structure data | EMDB-28650, PDB-8ex4: EMDB-28651, PDB-8ex5: EMDB-28652, PDB-8ex6: EMDB-28653, PDB-8ex7: EMDB-28654, PDB-8ex8: EMDB-29860, PDB-8g92: |
| Chemicals | 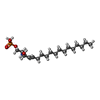 ChemComp-S1P: 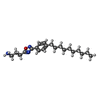 ChemComp-YUX: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / transporter / IMMUNE SYSTEM / SPNS2 / Sphingosine-1-phosphate / major facilitator superfamily |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human)