+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of inhibitor 16d-bound SPNS2 | |||||||||||||||
 Map data Map data | Structure of inhibitor 16d-bound SPNS2 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | SPNS2 / Sphingosine-1-phosphate / major facilitator superfamily / transporter / IMMUNE SYSTEM | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis ...regulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis / B cell homeostasis / transmembrane transporter activity / lymph node development / sensory perception of sound / bone development / endosome membrane / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||||||||
 Authors Authors | Chen H / Li X | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate. Authors: Hongwen Chen / Shahbaz Ahmed / Hongtu Zhao / Nadia Elghobashi-Meinhardt / Yaxin Dai / Jae Hun Kim / Jeffrey G McDonald / Xiaochun Li / Chia-Hsueh Lee /   Abstract: Sphingosine-1-phosphate (S1P) is an important signaling sphingolipid that regulates the immune system, angiogenesis, auditory function, and epithelial and endothelial barrier integrity. Spinster ...Sphingosine-1-phosphate (S1P) is an important signaling sphingolipid that regulates the immune system, angiogenesis, auditory function, and epithelial and endothelial barrier integrity. Spinster homolog 2 (Spns2) is an S1P transporter that exports S1P to initiate lipid signaling cascades. Modulating Spns2 activity can be beneficial in treatments of cancer, inflammation, and immune diseases. However, the transport mechanism of Spns2 and its inhibition remain unclear. Here, we present six cryo-EM structures of human Spns2 in lipid nanodiscs, including two functionally relevant intermediate conformations that link the inward- and outward-facing states, to reveal the structural basis of the S1P transport cycle. Functional analyses suggest that Spns2 exports S1P via facilitated diffusion, a mechanism distinct from other MFS lipid transporters. Finally, we show that the Spns2 inhibitor 16d attenuates the transport activity by locking Spns2 in the inward-facing state. Our work sheds light on Spns2-mediated S1P transport and aids the development of advanced Spns2 inhibitors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29860.map.gz emd_29860.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29860-v30.xml emd-29860-v30.xml emd-29860.xml emd-29860.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29860_fsc.xml emd_29860_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29860.png emd_29860.png | 47.1 KB | ||
| Filedesc metadata |  emd-29860.cif.gz emd-29860.cif.gz | 5.8 KB | ||
| Others |  emd_29860_half_map_1.map.gz emd_29860_half_map_1.map.gz emd_29860_half_map_2.map.gz emd_29860_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29860 http://ftp.pdbj.org/pub/emdb/structures/EMD-29860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29860 | HTTPS FTP |
-Related structure data
| Related structure data |  8g92MC  8ex4C  8ex5C  8ex6C  8ex7C  8ex8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29860.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29860.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of inhibitor 16d-bound SPNS2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_29860_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29860_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SPNS2 in complex with inhibitor 16d
| Entire | Name: SPNS2 in complex with inhibitor 16d |
|---|---|
| Components |
|
-Supramolecule #1: SPNS2 in complex with inhibitor 16d
| Supramolecule | Name: SPNS2 in complex with inhibitor 16d / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Sphingosine-1-phosphate transporter SPNS2
| Macromolecule | Name: Sphingosine-1-phosphate transporter SPNS2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.770148 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EKRKAEEEKR KAAAAILSLG NVLNYLDRYT VAGVLLDIQQ HFGVKDRGAG LLQSVFICSF MVAAPIFGYL GDRFNRKVIL SCGIFFWSA VTFSSSFIPQ QYFWLLVLSR GLVGIGEASY STIAPTIIGD LFTKNTRTLM LSVFYFAIPL GSGLGYITGS S VKQAAGDW ...String: EKRKAEEEKR KAAAAILSLG NVLNYLDRYT VAGVLLDIQQ HFGVKDRGAG LLQSVFICSF MVAAPIFGYL GDRFNRKVIL SCGIFFWSA VTFSSSFIPQ QYFWLLVLSR GLVGIGEASY STIAPTIIGD LFTKNTRTLM LSVFYFAIPL GSGLGYITGS S VKQAAGDW HWALRVSPVL GMITGTLILI LVPATKRGHA DQLGDQLKAR TSWLRDMKAL IRNRSYVFSS LATSAVSFAT GA LGMWIPL YLHRAQVVQK TAETCNSPPC GAKDSLIFGA ITCFTGFLGV VTGAGATRWC RLKTQRADPL VCAVGMLGSA IFI CLIFVA AKSSIVGAYI CIFVGETLLF SNWAITADIL MYVVIPTRRA TAVALQSFTS HLLGDAGSPY LIGFISDLIR QSTK DSPLW EFLSLGYALM LCPFVVVLGG MFFLATALFF VSDRARAEQQ VNQLAMPPAS VKV UniProtKB: Sphingosine-1-phosphate transporter SPNS2 |
-Macromolecule #2: 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine
| Macromolecule | Name: 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine type: ligand / ID: 2 / Number of copies: 1 / Formula: YUX |
|---|---|
| Molecular weight | Theoretical: 343.506 Da |
| Chemical component information | 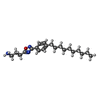 ChemComp-YUX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)