+Search query
-Structure paper
| Title | In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog. |
|---|---|
| Journal, issue, pages | Mol Microbiol, Vol. 114, Issue 3, Page 443-453, Year 2020 |
| Publish date | May 25, 2020 |
 Authors Authors | Florian M Rossmann / Isabelle Hug / Matteo Sangermani / Urs Jenal / Morgan Beeby /   |
| PubMed Abstract | Bacterial flagellar motility is controlled by the binding of CheY proteins to the cytoplasmic switch complex of the flagellar motor, resulting in changes in swimming speed or direction. Despite its ...Bacterial flagellar motility is controlled by the binding of CheY proteins to the cytoplasmic switch complex of the flagellar motor, resulting in changes in swimming speed or direction. Despite its importance for motor function, structural information about the interaction between effector proteins and the motor are scarce. To address this gap in knowledge, we used electron cryotomography and subtomogram averaging to visualize such interactions inside Caulobacter crescentus cells. In C. crescentus, several CheY homologs regulate motor function for different aspects of the bacterial lifestyle. We used subtomogram averaging to image binding of the CheY family protein CleD to the cytoplasmic Cring switch complex, the control center of the flagellar motor. This unambiguously confirmed the orientation of the motor switch protein FliM and the binding of a member of the CheY protein family to the outside rim of the C ring. We also uncovered previously unknown structural elaborations of the alphaproteobacterial flagellar motor, including two novel periplasmic ring structures, and the stator ring harboring eleven stator units, adding to our growing catalog of bacterial flagellar diversity. |
 External links External links |  Mol Microbiol / Mol Microbiol /  PubMed:32449846 / PubMed:32449846 /  PubMed Central PubMed Central |
| Methods | EM (subtomogram averaging) |
| Resolution | 82.0 - 110.0 Å |
| Structure data |  EMDB-10943:  EMDB-10945: 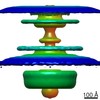 EMDB-10949:  EMDB-10950: 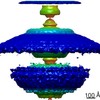 EMDB-10955:  EMDB-10956: 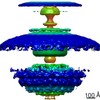 EMDB-10957: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Caulobacter vibrioides CB15 (bacteria)
Caulobacter vibrioides CB15 (bacteria)