[English] 日本語
 Yorodumi
Yorodumi- EMDB-10945: In situ structure of the Caulobacter crescentus flagellar motor a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10945 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog | |||||||||
 Map data Map data | Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs deletion strain, C11 symmetrised | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Caulobacter vibrioides CB15 (bacteria) Caulobacter vibrioides CB15 (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 82.0 Å | |||||||||
 Authors Authors | Rossmann FM / Hug I / Sangermani M / Jenal U / Beeby M | |||||||||
| Funding support |  Germany, Germany,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Microbiol / Year: 2020 Journal: Mol Microbiol / Year: 2020Title: In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog. Authors: Florian M Rossmann / Isabelle Hug / Matteo Sangermani / Urs Jenal / Morgan Beeby /   Abstract: Bacterial flagellar motility is controlled by the binding of CheY proteins to the cytoplasmic switch complex of the flagellar motor, resulting in changes in swimming speed or direction. Despite its ...Bacterial flagellar motility is controlled by the binding of CheY proteins to the cytoplasmic switch complex of the flagellar motor, resulting in changes in swimming speed or direction. Despite its importance for motor function, structural information about the interaction between effector proteins and the motor are scarce. To address this gap in knowledge, we used electron cryotomography and subtomogram averaging to visualize such interactions inside Caulobacter crescentus cells. In C. crescentus, several CheY homologs regulate motor function for different aspects of the bacterial lifestyle. We used subtomogram averaging to image binding of the CheY family protein CleD to the cytoplasmic Cring switch complex, the control center of the flagellar motor. This unambiguously confirmed the orientation of the motor switch protein FliM and the binding of a member of the CheY protein family to the outside rim of the C ring. We also uncovered previously unknown structural elaborations of the alphaproteobacterial flagellar motor, including two novel periplasmic ring structures, and the stator ring harboring eleven stator units, adding to our growing catalog of bacterial flagellar diversity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10945.map.gz emd_10945.map.gz | 54.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10945-v30.xml emd-10945-v30.xml emd-10945.xml emd-10945.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10945.png emd_10945.png | 68.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10945 http://ftp.pdbj.org/pub/emdb/structures/EMD-10945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10945 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10945.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10945.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs deletion strain, C11 symmetrised | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.00206 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs d...
| Entire | Name: Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs deletion strain, C11 symmetrised |
|---|---|
| Components |
|
-Supramolecule #1: Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs d...
| Supramolecule | Name: Flagellar motor of Caulobactor crescentus CB 15, in situ, cheYs deletion strain, C11 symmetrised type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides CB15 (bacteria) Caulobacter vibrioides CB15 (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Name: peptone yeast extract; PYE medium |
|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: blot time 5 s, blot force 3, wait time 60s, drain time 0.5s,. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 7 / Average exposure time: 1.5 sec. / Average electron dose: 3.15 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C11 (11 fold cyclic) / Algorithm: SIMULTANEOUS ITERATIVE (SIRT) / Resolution.type: BY AUTHOR / Resolution: 82.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET (ver. 1.10.1) / Number subtomograms used: 112 |
|---|---|
| Extraction | Number tomograms: 341 / Number images used: 278 / Reference model: sperical / Method: manual / Software - Name: PEET (ver. 1.10.1) |
| Final 3D classification | Software - Name: PEET (ver. 1.10.1) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: PEET (ver. 1.10.1) |
 Movie
Movie Controller
Controller




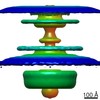

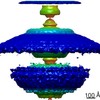

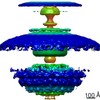




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















