+Search query
-Structure paper
| Title | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant. |
|---|---|
| Journal, issue, pages | Commun Biol, Vol. 4, Issue 1, Page 1380, Year 2021 |
| Publish date | Dec 9, 2021 |
 Authors Authors | Ido Caspy / Tom Schwartz / Vinzenz Bayro-Kaiser / Mariia Fadeeva / Amit Kessel / Nir Ben-Tal / Nathan Nelson /  |
| PubMed Abstract | Water molecules play a pivotal functional role in photosynthesis, primarily as the substrate for Photosystem II (PSII). However, their importance and contribution to Photosystem I (PSI) activity ...Water molecules play a pivotal functional role in photosynthesis, primarily as the substrate for Photosystem II (PSII). However, their importance and contribution to Photosystem I (PSI) activity remains obscure. Using a high-resolution cryogenic electron microscopy (cryo-EM) PSI structure from a Chlamydomonas reinhardtii temperature-sensitive photoautotrophic PSII mutant (TSP4), a conserved network of water molecules - dating back to cyanobacteria - was uncovered, mainly in the vicinity of the electron transport chain (ETC). The high-resolution structure illustrated that the water molecules served as a ligand in every chlorophyll that was missing a fifth magnesium coordination in the PSI core and in the light-harvesting complexes (LHC). The asymmetric distribution of the water molecules near the ETC branches modulated their electrostatic landscape, distinctly in the space between the quinones and FX. The data also disclosed the first observation of eukaryotic PSI oligomerisation through a low-resolution PSI dimer that was comprised of PSI-10LHC and PSI-8LHC. |
 External links External links |  Commun Biol / Commun Biol /  PubMed:34887518 / PubMed:34887518 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.54 - 17.1 Å |
| Structure data | EMDB-12180, PDB-7bgi: EMDB-12227, PDB-7blx: EMDB-12672, PDB-7o01: |
| Chemicals |  ChemComp-CL0:  ChemComp-CLA:  ChemComp-PQN:  ChemComp-SF4:  ChemComp-BCR:  ChemComp-LHG: 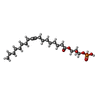 ChemComp-NKP:  ChemComp-LMT: 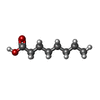 ChemComp-OCA:  ChemComp-DAO:  ChemComp-LMG:  ChemComp-DGA:  ChemComp-DGD:  ChemComp-CA:  ChemComp-T7X:  ChemComp-RRX: 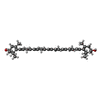 ChemComp-C7Z:  ChemComp-SPH:  ChemComp-LUT:  ChemComp-CHL:  ChemComp-SQD:  ChemComp-QTB:  ChemComp-3PH:  ChemComp-PLM: 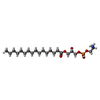 ChemComp-LPX:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  PHOTOSYNTHESIS / PHOTOSYNTHESIS /  chlamydomonas / chlamydomonas /  photosystem I / temperature sensitive / photosystem I / temperature sensitive /  water molecules water molecules |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers










