+Search query
-Structure paper
| Title | Cryo-EM of human P-glycoprotein reveals an intermediate occluded conformation during active drug transport. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 16, Issue 1, Page 3619, Year 2025 |
| Publish date | Apr 16, 2025 |
 Authors Authors | Alan T Culbertson / Maofu Liao /   |
| PubMed Abstract | P-glycoprotein (Pgp) is an important human multidrug transporter that contributes to pharmacokinetics and multidrug resistance. Despite decades of study, the conformation transition cycle of Pgp ...P-glycoprotein (Pgp) is an important human multidrug transporter that contributes to pharmacokinetics and multidrug resistance. Despite decades of study, the conformation transition cycle of Pgp undergoing active drug transport is not defined, thus the precise relevance of all available Pgp structures to uninterrupted multidrug transport remains unclear. Here, we use cryo-EM of membrane-embedded human Pgp under continuous turnover conditions to analyze the conformational ensembles of Pgp transporting distinct substrates. These results delineate multiple conformations including inward-facing and closed conformations, highlighting the occluded conformation as a critical intermediate state between transporter closure and substrate release. A combination of structural, functional, and computational studies reveals the transmembrane helices 4 and 10 undergoing drastic rearrangement to coordinate substrate binding, occlusion, and release, and identifies a peripheral site involved in substrate capture and Pgp inhibition. Together, our results provide a set of snapshots of Pgp undergoing continuous drug transport, unveiling the intricate interplay between transporter dynamics and drug movement, and shed light on the mechanism of polyspecificity. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:40240353 / PubMed:40240353 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.9 - 6.4 Å |
| Structure data | EMDB-40226, PDB-8gmg: EMDB-40227, PDB-8gmj: EMDB-40258, PDB-8sa0: EMDB-40259, PDB-8sa1: EMDB-40292, PDB-8sb7: EMDB-40293, PDB-8sb8: EMDB-40294, PDB-8sb9: EMDB-40295, PDB-8sba:  EMDB-40326: CryoEM structure of P-Glycoprotein in inward facing state with MgATP and vanadate  EMDB-40340: CryoEM structure of P-Glycoprotein in inward facing state 1 with vinblastine  EMDB-40341: CryoEM structure of P-Glycoprotein in inward facing state 2 with vinblastine  EMDB-40342: CryoEM structure of P-Glycoprotein in closed 1 state under continuous turnover conditions with vinblastine  EMDB-40343: CryoEM structure of P-Glycoprotein in closed 2 state under continuous turnover conditions with vinblastine |
| Chemicals |  ChemComp-MG:  ChemComp-ATP: 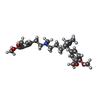 ChemComp-I6H: 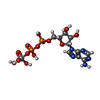 ChemComp-AOV: 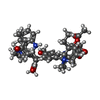 ChemComp-VLB: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / multidrug resistance / ABC transporter / membrane protein / transporter / Transporter. |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



















 homo sapiens (human)
homo sapiens (human)