+Search query
-Structure paper
| Title | Structural basis for the reversal of human MRP4-mediated multidrug resistance by lapatinib. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 44, Issue 4, Page 115466, Year 2025 |
| Publish date | Mar 25, 2025 |
 Authors Authors | Zhipeng Xie / Jiaxiang Lv / Wei Huang / Zhikun Wu / Rongli Zhu / Zixin Deng / Feng Long /  |
| PubMed Abstract | Multidrug resistance proteins (MRPs) are one of the major mechanisms for developing cancer drug resistance. Human MRP4 (hMRP4) plays an important role in various chemotherapy-resistant cancers. Here, ...Multidrug resistance proteins (MRPs) are one of the major mechanisms for developing cancer drug resistance. Human MRP4 (hMRP4) plays an important role in various chemotherapy-resistant cancers. Here, we show hMRP4 mediates the resistance of a broad spectrum of antitumor reagents in the cultured tumor cells, among which the cell resistance to vincristine and 5-fluorouracil is rescued by supplementing a tyrosinase inhibitor, lapatinib. The cryoelectron microscopy (cryo-EM) structures of hMRP4 in the substrate- or inhibitor-bound form are determined. Although lapatinib shares partial binding sites with vincristine and 5-fluorouracil using a similar set of crucial residues located in the central cavity of hMRP4, the high binding affinity of lapatinib and its unique binding mode with transmembrane helices TM2 and TM12 inside the pathway tunnel prohibit hMRP4 from structural transition between intermediate states during drug translocation. This study provides mechanistic insights into the therapeutical potential of lapatinib in combating hMRP4-mediated MDR. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:40138312 PubMed:40138312 |
| Methods | EM (single particle) |
| Resolution | 2.96 - 3.9 Å |
| Structure data | EMDB-39909, PDB-8zbs: EMDB-39910, PDB-8zbt: EMDB-39911, PDB-8zbu: |
| Chemicals |  ChemComp-R1Q:  ChemComp-ANP:  ChemComp-MG: 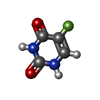 ChemComp-URF:  ChemComp-ATP:  ChemComp-FMM: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / ATP-binding cassette sub-family C member 4 / Multidrug resistance proteins (MRPs) / multidrug-resistance (MDR) / TRANSPORT PROTEIN |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)