+Search query
-Structure paper
| Title | Two riboswitch classes that share a common ligand-binding fold show major differences in the ability to accommodate mutations. |
|---|---|
| Journal, issue, pages | Nucleic Acids Res., Vol. 52, Page 13152-13173, Year 2024 |
| Publish date | Jun 26, 2020 (structure data deposition date) |
 Authors Authors | Srivastava, Y. / Akinyemi, O. / Rohe, T.C. / Pritchett, E.M. / Baker, C.D. / Sharma, A. / Jenkins, J.L. / Mathews, D.H. / Wedekind, J.E. |
 External links External links |  Nucleic Acids Res. / Nucleic Acids Res. /  PubMed:39413212 PubMed:39413212 |
| Methods | X-ray diffraction |
| Resolution | 2.73 - 3.04 Å |
| Structure data |  PDB-6xkn:  PDB-6xko:  PDB-8toz:  PDB-8vpv: |
| Chemicals | 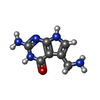 ChemComp-PRF:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | RNA / PREQ1 / QUEUOSINE / THREE-WAY HELICAL JUNCTION / APTAMER / METABOLITE / TRANSLATIONAL REGULATION / HL(OUT)-TYPE PSEUDOKNOT / RIBOSWITCH |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 faecalibacterium prausnitzii (bacteria)
faecalibacterium prausnitzii (bacteria)