+Search query
-Structure paper
| Title | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 10, Issue 31, Page eadp2211, Year 2024 |
| Publish date | Aug 2, 2024 |
 Authors Authors | Ying Yin / Cheon-Gyu Park / Feng Zhang / Justin G Fedor / Shasha Feng / Yang Suo / Wonpil Im / Seok-Yong Lee /  |
| PubMed Abstract | Our sensory adaptation to cold and chemically induced coolness is mediated by the intrinsic property of TRPM8 channels to desensitize. TRPM8 is also implicated in cold-evoked pain disorders and ...Our sensory adaptation to cold and chemically induced coolness is mediated by the intrinsic property of TRPM8 channels to desensitize. TRPM8 is also implicated in cold-evoked pain disorders and migraine, highlighting its inhibitors as an avenue for pain relief. Despite the importance, the mechanisms of TRPM8 desensitization and inhibition remained unclear. We found, using cryo-electron microscopy, electrophysiology, and molecular dynamics simulations, that TRPM8 inhibitors bind selectively to the desensitized state of the channel. These inhibitors were used to reveal the overlapping mechanisms of desensitization and inhibition and that cold and cooling agonists share a common desensitization pathway. Furthermore, we identified the structural determinants crucial for the conformational change in TRPM8 desensitization. Our study illustrates how receptor-level conformational changes alter cold sensation, providing insights into therapeutic development. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:39093967 / PubMed:39093967 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.76 - 4.13 Å |
| Structure data | EMDB-44255, PDB-9b6d: EMDB-44256, PDB-9b6e: EMDB-44257, PDB-9b6f: EMDB-44258, PDB-9b6g: EMDB-44259, PDB-9b6h: EMDB-44260, PDB-9b6i: EMDB-44261, PDB-9b6j: EMDB-44262, PDB-9b6k: |
| Chemicals |  ChemComp-Y01: 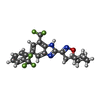 ChemComp-T14:  PDB-1aiy: 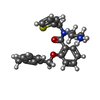 ChemComp-LQ7:  ChemComp-NAG:  ChemComp-ULO:  ChemComp-CA:  ChemComp-PIO: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / TRPM8 / menthol receptor / cold receptor / PI(4 / 5)P2 / cooling agonists / temperature sensing / ion channel / sensory transduction / transient receptor potential ion channel / TRPM8 activation / TRPM8 inhibition / TRPM8 desensitization / TRPM8 antagonists |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




















 parus major (Great Tit)
parus major (Great Tit)