[English] 日本語
 Yorodumi
Yorodumi- PDB-9b6e: Cryo-EM structure of the mouse TRPM8 channel in complex with the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9b6e | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 | |||||||||
 Components Components | Transient receptor potential cation channel subfamily M member 8 | |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / TRPM8 / menthol receptor / cold receptor / PI(4 / 5)P2 / cooling agonists / temperature sensing / ion channel / sensory transduction / transient receptor potential ion channel / TRPM8 activation / TRPM8 inhibition / TRPM8 desensitization / TRPM8 antagonists | |||||||||
| Function / homology |  Function and homology information Function and homology informationTRP channels / thermoception / response to temperature stimulus / monoatomic ion channel activity / plasma membrane raft / response to cold / calcium channel activity / intracellular calcium ion homeostasis / calcium ion transport / positive regulation of cold-induced thermogenesis ...TRP channels / thermoception / response to temperature stimulus / monoatomic ion channel activity / plasma membrane raft / response to cold / calcium channel activity / intracellular calcium ion homeostasis / calcium ion transport / positive regulation of cold-induced thermogenesis / membrane raft / external side of plasma membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Yin, Y. / Park, C.-G. / Zhang, F. / Fedor, J. / Feng, S. / Suo, Y. / Im, W. / Lee, S.-Y. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8. Authors: Ying Yin / Cheon-Gyu Park / Feng Zhang / Justin G Fedor / Shasha Feng / Yang Suo / Wonpil Im / Seok-Yong Lee /  Abstract: Our sensory adaptation to cold and chemically induced coolness is mediated by the intrinsic property of TRPM8 channels to desensitize. TRPM8 is also implicated in cold-evoked pain disorders and ...Our sensory adaptation to cold and chemically induced coolness is mediated by the intrinsic property of TRPM8 channels to desensitize. TRPM8 is also implicated in cold-evoked pain disorders and migraine, highlighting its inhibitors as an avenue for pain relief. Despite the importance, the mechanisms of TRPM8 desensitization and inhibition remained unclear. We found, using cryo-electron microscopy, electrophysiology, and molecular dynamics simulations, that TRPM8 inhibitors bind selectively to the desensitized state of the channel. These inhibitors were used to reveal the overlapping mechanisms of desensitization and inhibition and that cold and cooling agonists share a common desensitization pathway. Furthermore, we identified the structural determinants crucial for the conformational change in TRPM8 desensitization. Our study illustrates how receptor-level conformational changes alter cold sensation, providing insights into therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9b6e.cif.gz 9b6e.cif.gz | 1.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9b6e.ent.gz pdb9b6e.ent.gz | 1.1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9b6e.json.gz 9b6e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b6/9b6e https://data.pdbj.org/pub/pdb/validation_reports/b6/9b6e ftp://data.pdbj.org/pub/pdb/validation_reports/b6/9b6e ftp://data.pdbj.org/pub/pdb/validation_reports/b6/9b6e | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  44256MC  9b6dC  9b6fC  9b6gC  9b6hC  9b6iC  9b6jC  9b6kC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
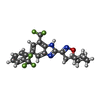
| #1: Protein | Mass: 131548.312 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q8R4D5 Homo sapiens (human) / References: UniProt: Q8R4D5#2: Chemical | ChemComp-T14 / #3: Chemical | ChemComp-Y01 / Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Transient receptor potential cation channel subfamily M member 8 Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 8 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 293.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Nominal defocus max: 2200 nm / Nominal defocus min: 700 nm / Cs: 2.7 mm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 9581 |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2896859 | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C4 (4 fold cyclic) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.91 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 194290 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||
| Atomic model building | Details: Another structural model from this study was used as the initial model for model building. Source name: Other / Type: experimental model | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj