+Search query
-Structure paper
| Title | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 40, Issue 4, Page 111133, Year 2022 |
| Publish date | Jul 26, 2022 |
 Authors Authors | Kuo Zhang / Michelle Cheok Yien Law / Trinh Mai Nguyen / Yaw Bia Tan / Melissa Wirawan / Yee-Song Law / Lak Shin Jeong / Dahai Luo /    |
| PubMed Abstract | Many viruses encode RNA-modifying enzymes to edit the 5' end of viral RNA to mimic the cellular mRNA for effective protein translation, genome replication, and evasion of the host defense mechanisms. ...Many viruses encode RNA-modifying enzymes to edit the 5' end of viral RNA to mimic the cellular mRNA for effective protein translation, genome replication, and evasion of the host defense mechanisms. Alphavirus nsP1 synthesizes the 5' end Cap-0 structure of viral RNAs. However, the molecular basis of the capping process remains unclear. We determine high-resolution cryoelectron microscopy (cryo-EM) structures of Chikungunya virus nsP1 in complex with m7GTP/SAH, covalently attached m7GMP, and Cap-0 viral RNA. These structures reveal details of viral-RNA-capping reactions and uncover a sequence-specific virus RNA-recognition pattern that, in turn, regulates viral-RNA-capping efficiency to ensure optimal genome replication and subgenomic RNA transcription. This sequence-specific enzyme-RNA pairing is conserved across all alphaviruses. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:35905713 PubMed:35905713 |
| Methods | EM (single particle) |
| Resolution | 2.18 - 2.62 Å |
| Structure data | EMDB-31580, PDB-7fgg: EMDB-31581, PDB-7fgh: EMDB-31582, PDB-7fgi: EMDB-32914, PDB-7x01: |
| Chemicals |  ChemComp-ZN:  ChemComp-SAH: 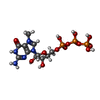 ChemComp-MGP:  ChemComp-MG:  ChemComp-HOH:  ChemComp-G7M: 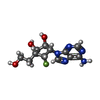 ChemComp-7XQ: |
| Source |
|
 Keywords Keywords | VIRAL PROTEIN / nonstructural protein / Chikungunya virus / RNA cap / replication / VIRAL PROTEIN/RNA / VIRAL PROTEIN-RNA complex / inhibitor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 chikungunya virus strain s27-african prototype
chikungunya virus strain s27-african prototype