[English] 日本語
 Yorodumi
Yorodumi- EMDB-31580: Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with m7GTP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nonstructural protein / Chikungunya virus / RNA cap / replication / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationADP-ribose 1''-phosphate phosphatase / host cell filopodium / mRNA methyltransferase activity / polynucleotide 5'-phosphatase activity / mRNA 5'-phosphatase / mRNA 5'-triphosphate monophosphatase activity / polynucleotide adenylyltransferase / poly(A) RNA polymerase activity / mRNA modification / regulation of cytoskeleton organization ...ADP-ribose 1''-phosphate phosphatase / host cell filopodium / mRNA methyltransferase activity / polynucleotide 5'-phosphatase activity / mRNA 5'-phosphatase / mRNA 5'-triphosphate monophosphatase activity / polynucleotide adenylyltransferase / poly(A) RNA polymerase activity / mRNA modification / regulation of cytoskeleton organization / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / cysteine-type peptidase activity / Transferases; Transferring one-carbon groups; Methyltransferases / host cell cytoplasmic vesicle membrane / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / nucleoside-triphosphate phosphatase / methylation / RNA helicase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / symbiont-mediated suppression of host innate immune response / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated suppression of host gene expression / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / GTP binding / host cell nucleus / host cell plasma membrane / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.19 Å | |||||||||
 Authors Authors | Zhang K / Law MCY | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1. Authors: Kuo Zhang / Michelle Cheok Yien Law / Trinh Mai Nguyen / Yaw Bia Tan / Melissa Wirawan / Yee-Song Law / Lak Shin Jeong / Dahai Luo /    Abstract: Many viruses encode RNA-modifying enzymes to edit the 5' end of viral RNA to mimic the cellular mRNA for effective protein translation, genome replication, and evasion of the host defense mechanisms. ...Many viruses encode RNA-modifying enzymes to edit the 5' end of viral RNA to mimic the cellular mRNA for effective protein translation, genome replication, and evasion of the host defense mechanisms. Alphavirus nsP1 synthesizes the 5' end Cap-0 structure of viral RNAs. However, the molecular basis of the capping process remains unclear. We determine high-resolution cryoelectron microscopy (cryo-EM) structures of Chikungunya virus nsP1 in complex with m7GTP/SAH, covalently attached m7GMP, and Cap-0 viral RNA. These structures reveal details of viral-RNA-capping reactions and uncover a sequence-specific virus RNA-recognition pattern that, in turn, regulates viral-RNA-capping efficiency to ensure optimal genome replication and subgenomic RNA transcription. This sequence-specific enzyme-RNA pairing is conserved across all alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31580.map.gz emd_31580.map.gz | 398.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31580-v30.xml emd-31580-v30.xml emd-31580.xml emd-31580.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31580.png emd_31580.png | 68.6 KB | ||
| Filedesc metadata |  emd-31580.cif.gz emd-31580.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31580 http://ftp.pdbj.org/pub/emdb/structures/EMD-31580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31580 | HTTPS FTP |
-Related structure data
| Related structure data |  7fggMC  7fghC  7fgiC  7x01C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31580.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31580.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Nonstructural Protein 1
| Entire | Name: Nonstructural Protein 1 |
|---|---|
| Components |
|
-Supramolecule #1: Nonstructural Protein 1
| Supramolecule | Name: Nonstructural Protein 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
-Macromolecule #1: mRNA-capping enzyme nsP1
| Macromolecule | Name: mRNA-capping enzyme nsP1 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain S27-African prototype Chikungunya virus strain S27-African prototype |
| Molecular weight | Theoretical: 62.056539 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDPVYVDIDA DSAFLKALQR AYPMFEVEPR QVTPNDHANA RAFSHLAIKL IEQEIDPDST ILDIGSAPAR RMMSDRKYHC VCPMRSAED PERLANYARK LASAAGKVLD RNISGKIGDL QAVMAVPDTE TPTFCLHTDV SCRQRADVAI YQDVYAVHAP T SLYHQAIK ...String: MDPVYVDIDA DSAFLKALQR AYPMFEVEPR QVTPNDHANA RAFSHLAIKL IEQEIDPDST ILDIGSAPAR RMMSDRKYHC VCPMRSAED PERLANYARK LASAAGKVLD RNISGKIGDL QAVMAVPDTE TPTFCLHTDV SCRQRADVAI YQDVYAVHAP T SLYHQAIK GVRLAYWVGF DTTPFMYNAM AGAYPSYSTN WADEQVLKAK NIGLCSTDLT EGRRGKLSIM RGKKLEPCDR VL FSVGSTL YPESRKLLKS WHLPSVFHLK GKLSFTCRCD TVVSCEGYVV KRITMSPGLY GKTTGYAVTH HADGFLMCKT TDT VDGERV SFSVCTYVPA TICDQMTGIL ATEVTPEDAQ KLLVGLNQRI VVNGRTQRNT NTMKNYMIPV VAQAFSKWAK ECRK DMEDE KLLGVRERTL TCCCLWAFKK QKTHTVYKRP DTQSIQKVQA EFDSFVVPSL WSSGLSIPLR TRIKWLLSKV PKTDL TPYS GDAQEARDAE KEAEEEREAE LTLEALPPLQ AAGGGGSWSH PQFEKMDYKD HDGDYKDHDI DYKDDDDK UniProtKB: Polyprotein P1234 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 12 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: S-ADENOSYL-L-HOMOCYSTEINE
| Macromolecule | Name: S-ADENOSYL-L-HOMOCYSTEINE / type: ligand / ID: 3 / Number of copies: 12 / Formula: SAH |
|---|---|
| Molecular weight | Theoretical: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
-Macromolecule #4: 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 12 / Formula: MGP |
|---|---|
| Molecular weight | Theoretical: 538.215 Da |
| Chemical component information | 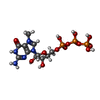 ChemComp-MGP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 1764 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.19 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 244484 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)