+Search query
-Structure paper
| Title | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 12, Issue 1, Page 4223, Year 2021 |
| Publish date | Jul 9, 2021 |
 Authors Authors | Akihiro Kawamoto / Tomoko Miyata / Fumiaki Makino / Miki Kinoshita / Tohru Minamino / Katsumi Imada / Takayuki Kato / Keiichi Namba /  |
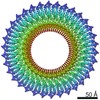
| PubMed Abstract | The bacterial flagellar MS ring is a transmembrane complex acting as the core of the flagellar motor and template for flagellar assembly. The C ring attached to the MS ring is involved in torque ...The bacterial flagellar MS ring is a transmembrane complex acting as the core of the flagellar motor and template for flagellar assembly. The C ring attached to the MS ring is involved in torque generation and rotation switch, and a large symmetry mismatch between these two rings has been a long puzzle, especially with respect to their role in motor function. Here, using cryoEM structural analysis of the flagellar basal body and the MS ring formed by full-length FliF from Salmonella enterica, we show that the native MS ring is formed by 34 FliF subunits with no symmetry variation. Symmetry analysis of the C ring shows a variation with a peak at 34-fold, suggesting flexibility in C ring assembly. Finally, our data also indicate that FliF subunits assume two different conformations, contributing differentially to the inner and middle parts of the M ring and thus resulting in 23- and 11-fold subsymmetries in the inner and middle M ring, respectively. The internal core of the M ring, formed by 23 subunits, forms a hole of the right size to accommodate the protein export gate. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:34244518 / PubMed:34244518 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 11.6 Å |
| Structure data |  EMDB-30360:  EMDB-30361:  EMDB-30363: EMDB-30612, PDB-7d84:  EMDB-30613:  EMDB-30940:  EMDB-30941:  EMDB-30942: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Flagellar motor / Type III protein export system / Transmembrane protein Complex / Torque generation |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)
Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)