+Search query
-Structure paper
| Title | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets. |
|---|---|
| Journal, issue, pages | J Am Chem Soc, Vol. 141, Issue 2, Page 936-944, Year 2019 |
| Publish date | Jan 16, 2019 |
 Authors Authors | Maximilian M Sauer / Roman P Jakob / Thomas Luber / Fabia Canonica / Giulio Navarra / Beat Ernst / Carlo Unverzagt / Timm Maier / Rudi Glockshuber /   |
| PubMed Abstract | Multivalent carbohydrate-lectin interactions at host-pathogen interfaces play a crucial role in the establishment of infections. Although competitive antagonists that prevent pathogen adhesion are ...Multivalent carbohydrate-lectin interactions at host-pathogen interfaces play a crucial role in the establishment of infections. Although competitive antagonists that prevent pathogen adhesion are promising antimicrobial drugs, the molecular mechanisms underlying these complex adhesion processes are still poorly understood. Here, we characterize the interactions between the fimbrial adhesin FimH from uropathogenic Escherichia coli strains and its natural high-mannose type N-glycan binding epitopes on uroepithelial glycoproteins. Crystal structures and a detailed kinetic characterization of ligand-binding and dissociation revealed that the binding pocket of FimH evolved such that it recognizes the terminal α(1-2)-, α(1-3)-, and α(1-6)-linked mannosides of natural high-mannose type N-glycans with similar affinity. We demonstrate that the 2000-fold higher affinity of the domain-separated state of FimH compared to its domain-associated state is ligand-independent and consistent with a thermodynamic cycle in which ligand-binding shifts the association equilibrium between the FimH lectin and the FimH pilin domain. Moreover, we show that a single N-glycan can bind up to three molecules of FimH, albeit with negative cooperativity, so that a molar excess of accessible N-glycans over FimH on the cell surface favors monovalent FimH binding. Our data provide pivotal insights into the adhesion properties of uropathogenic Escherichia coli strains to their target receptors and a solid basis for the development of effective FimH antagonists. |
 External links External links |  J Am Chem Soc / J Am Chem Soc /  PubMed:30543411 PubMed:30543411 |
| Methods | X-ray diffraction |
| Resolution | 1.719 - 2.501 Å |
| Structure data |  PDB-6gtv:  PDB-6gtw: 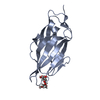 PDB-6gtx: 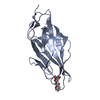 PDB-6gty: 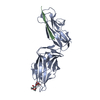 PDB-6gtz:  PDB-6gu0: |
| Chemicals |  ChemComp-PEG:  ChemComp-1PE:  ChemComp-HOH:  ChemComp-CA:  ChemComp-SO4: |
| Source |
|
 Keywords Keywords | CELL ADHESION / TYPE I PILUS / CATCH-BOND / LECTIN / UPEC / INFECTION / MANNOSE |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




