+Search query
-Structure paper
| Title | Structure of the human dopamine transporter and mechanisms of inhibition. |
|---|---|
| Journal, issue, pages | Nature, Vol. 632, Issue 8025, Page 672-677, Year 2024 |
| Publish date | Aug 7, 2024 |
 Authors Authors | Dushyant Kumar Srivastava / Vikas Navratna / Dilip K Tosh / Audrey Chinn / Md Fulbabu Sk / Emad Tajkhorshid / Kenneth A Jacobson / Eric Gouaux /  |
| PubMed Abstract | The neurotransmitter dopamine has central roles in mood, appetite, arousal and movement. Despite its importance in brain physiology and function, and as a target for illicit and therapeutic drugs, ...The neurotransmitter dopamine has central roles in mood, appetite, arousal and movement. Despite its importance in brain physiology and function, and as a target for illicit and therapeutic drugs, the human dopamine transporter (hDAT) and mechanisms by which it is inhibited by small molecules and Zn are without a high-resolution structural context. Here we determine the structure of hDAT in a tripartite complex with the competitive inhibitor and cocaine analogue, (-)-2-β-carbomethoxy-3-β-(4-fluorophenyl)tropane (β-CFT), the non-competitive inhibitor MRS7292 and Zn (ref. ). We show how β-CFT occupies the central site, approximately halfway across the membrane, stabilizing the transporter in an outward-open conformation. MRS7292 binds to a structurally uncharacterized allosteric site, adjacent to the extracellular vestibule, sequestered underneath the extracellular loop 4 (EL4) and adjacent to transmembrane helix 1b (TM1b), acting as a wedge, precluding movement of TM1b and closure of the extracellular gate. A Zn ion further stabilizes the outward-facing conformation by coupling EL4 to EL2, TM7 and TM8, thus providing specific insights into how Zn restrains the movement of EL4 relative to EL2 and inhibits transport activity. |
 External links External links |  Nature / Nature /  PubMed:39112705 / PubMed:39112705 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.19 Å |
| Structure data | EMDB-43128, PDB-8vby: |
| Chemicals | 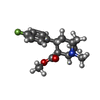 ChemComp-42L:  PDB-1aaf:  ChemComp-NAG:  ChemComp-Y01:  ChemComp-D10:  ChemComp-LNK:  ChemComp-HP6:  ChemComp-ZN:  ChemComp-NA:  ChemComp-D12:  ChemComp-MYS:  ChemComp-OCT:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Transport / Membrane / Inhibitor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)