[English] 日本語
 Yorodumi
Yorodumi- PDB-8vby: Structure of the human dopamine transporter in complex with beta-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8vby | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human dopamine transporter in complex with beta-CFT, MRS7292 and divalent zinc | |||||||||
 Components Components | Sodium-dependent dopamine transporter | |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Transport / Membrane / Inhibitor | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Dopamine clearance from the synaptic cleft / amine binding / dopamine uptake / adenohypophysis development / hyaloid vascular plexus regression / dopamine binding / norepinephrine:sodium symporter activity / dopamine:sodium symporter activity ...Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Dopamine clearance from the synaptic cleft / amine binding / dopamine uptake / adenohypophysis development / hyaloid vascular plexus regression / dopamine binding / norepinephrine:sodium symporter activity / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / regulation of dopamine metabolic process / dopamine transport / flotillin complex / dopamine catabolic process / dopaminergic synapse / monoamine transmembrane transporter activity / monoamine transport / positive regulation of multicellular organism growth / heterocyclic compound binding / SLC-mediated transport of neurotransmitters / response to iron ion / dopamine biosynthetic process / neurotransmitter transport / dopamine uptake involved in synaptic transmission / amino acid transport / response to cAMP / prepulse inhibition / lactation / axon terminus / protein phosphatase 2A binding / sodium ion transmembrane transport / response to nicotine / response to cocaine / locomotory behavior / cognition / sensory perception of smell / presynaptic membrane / protease binding / response to ethanol / postsynaptic membrane / neuron projection / membrane raft / response to xenobiotic stimulus / signaling receptor binding / axon / neuronal cell body / protein-containing complex binding / cell surface / metal ion binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Srivastava, D.K. / Gouaux, E. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structure of the human dopamine transporter and mechanisms of inhibition. Authors: Dushyant Kumar Srivastava / Vikas Navratna / Dilip K Tosh / Audrey Chinn / Md Fulbabu Sk / Emad Tajkhorshid / Kenneth A Jacobson / Eric Gouaux /  Abstract: The neurotransmitter dopamine has central roles in mood, appetite, arousal and movement. Despite its importance in brain physiology and function, and as a target for illicit and therapeutic drugs, ...The neurotransmitter dopamine has central roles in mood, appetite, arousal and movement. Despite its importance in brain physiology and function, and as a target for illicit and therapeutic drugs, the human dopamine transporter (hDAT) and mechanisms by which it is inhibited by small molecules and Zn are without a high-resolution structural context. Here we determine the structure of hDAT in a tripartite complex with the competitive inhibitor and cocaine analogue, (-)-2-β-carbomethoxy-3-β-(4-fluorophenyl)tropane (β-CFT), the non-competitive inhibitor MRS7292 and Zn (ref. ). We show how β-CFT occupies the central site, approximately halfway across the membrane, stabilizing the transporter in an outward-open conformation. MRS7292 binds to a structurally uncharacterized allosteric site, adjacent to the extracellular vestibule, sequestered underneath the extracellular loop 4 (EL4) and adjacent to transmembrane helix 1b (TM1b), acting as a wedge, precluding movement of TM1b and closure of the extracellular gate. A Zn ion further stabilizes the outward-facing conformation by coupling EL4 to EL2, TM7 and TM8, thus providing specific insights into how Zn restrains the movement of EL4 relative to EL2 and inhibits transport activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8vby.cif.gz 8vby.cif.gz | 168.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8vby.ent.gz pdb8vby.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8vby.json.gz 8vby.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vb/8vby https://data.pdbj.org/pub/pdb/validation_reports/vb/8vby ftp://data.pdbj.org/pub/pdb/validation_reports/vb/8vby ftp://data.pdbj.org/pub/pdb/validation_reports/vb/8vby | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  43128MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein / Sugars , 2 types, 2 molecules A

| #1: Protein | Mass: 63384.500 Da / Num. of mol.: 1 / Mutation: I248Y Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC6A3 / Production host: Homo sapiens (human) / Gene: SLC6A3 / Production host:  Homo sapiens (human) / References: UniProt: Q01959 Homo sapiens (human) / References: UniProt: Q01959 |
|---|---|
| #4: Sugar | ChemComp-NAG / |
-Non-polymers , 12 types, 50 molecules 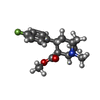


















| #2: Chemical | ChemComp-42L / | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-A1AAF / Mass: 459.906 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C20H18ClN5O4S / Feature type: SUBJECT OF INVESTIGATION | ||||||||||||||||||
| #5: Chemical | ChemComp-Y01 / #6: Chemical | ChemComp-D10 / #7: Chemical | ChemComp-LNK / #8: Chemical | ChemComp-HP6 / #9: Chemical | ChemComp-ZN / | #10: Chemical | ChemComp-NA / | #11: Chemical | ChemComp-D12 / #12: Chemical | ChemComp-MYS / | #13: Chemical | #14: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Sodium dependent dopamine transporter in complex with inhibitor Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.063 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||
| Buffer solution | pH: 7.8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: monodisperse sample | ||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 288.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 165000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
| EM imaging optics | Energyfilter name: TFS Selectris X / Energyfilter slit width: 6 eV |
- Processing
Processing
| EM software | Name: cryoSPARC / Category: CTF correction | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Image processing | Details: Falcon 4i | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.19 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 177494 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 53.23 Å2 | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj















