-Search query
-Search result
Showing all 35 items for (author: wong & ccl)

EMDB-34935: 
Cryo-EM structure of a SIN3/HDAC complex from budding yeast
Method: single particle / : Guo Z, Zhan X, Wang C
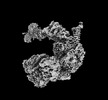
EMDB-34936: 
Cryo-EM map of the Rpd3L complex from the Rxt1-Flag dataset
Method: single particle / : Guo Z, Zhan X, Wang C

EMDB-34937: 
Cryo-EM map of the Rpd3L complex in Sin3 part
Method: single particle / : Guo Z, Zhan X, Wang C

EMDB-34938: 
Cryo-EM map of the Rpd3L complex in Dep1 region
Method: single particle / : Guo Z, Zhan X, Wang C
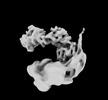
EMDB-34939: 
Cryo-EM map of the Rpd3L complex in Ume1 part
Method: single particle / : Guo Z, Zhan X, Wang C

EMDB-34227: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 1
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34229: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 4
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34230: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 2
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34235: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 3
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34236: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 5
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF
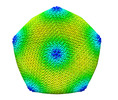
EMDB-34251: 
Singapore Grouper Iridovirus
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34815: 
One asymmetric unit of Singapore grouper iridovirus capsid
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-20238: 
Dimeric structure of ACAT1
Method: single particle / : Yan N, Qian HW

EMDB-20239: 
Tetrameric structure of ACAT1
Method: single particle / : Yan N, Qian HW

EMDB-9769: 
Yeast proteasome in translocation competent state (C3-a)
Method: single particle / : Cong Y

EMDB-6913: 
4.9 Angstrom Cryo-EM structure of human mTOR complex 2
Method: single particle / : Chen X, Liu M, Tian Y, Wang H, Wang J, Xu Y

EMDB-6668: 
Cryo-EM structure of mTORC1
Method: single particle / : Yang H, Wang J, Liu M, Chen X, Huang M, Tan D, Dong M, Wong CCL, Xu Y, Wang H

EMDB-6561: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP at 3.8 A
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6562: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for U4/U6 and U5 RNA region at 3.75 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6563: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for U4/U6 RNA region at 3.57 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6564: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for Prp31 region at 3.52 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6565: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for Prp8 region at 3.49 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6566: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for Prp3 region at 3.46 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6567: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for Prp4 region at 3.61 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6568: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for Prp4 WD40 region at 3.70 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6569: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for U5 RNA region at 3.70 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6570: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for U5 Snu114 region at 3.77 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y

EMDB-6571: 
The cryo-EM structure of yeast spliceosomal U4/U6.U5 tri-snRNP (improved map for U5 Sm ring region at 4.7 A)
Method: single particle / : Wan R, Yan C, Bai R, Wang L, Huang M, Wong C, Shi Y
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search










 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
